HorA Sever Tutorial
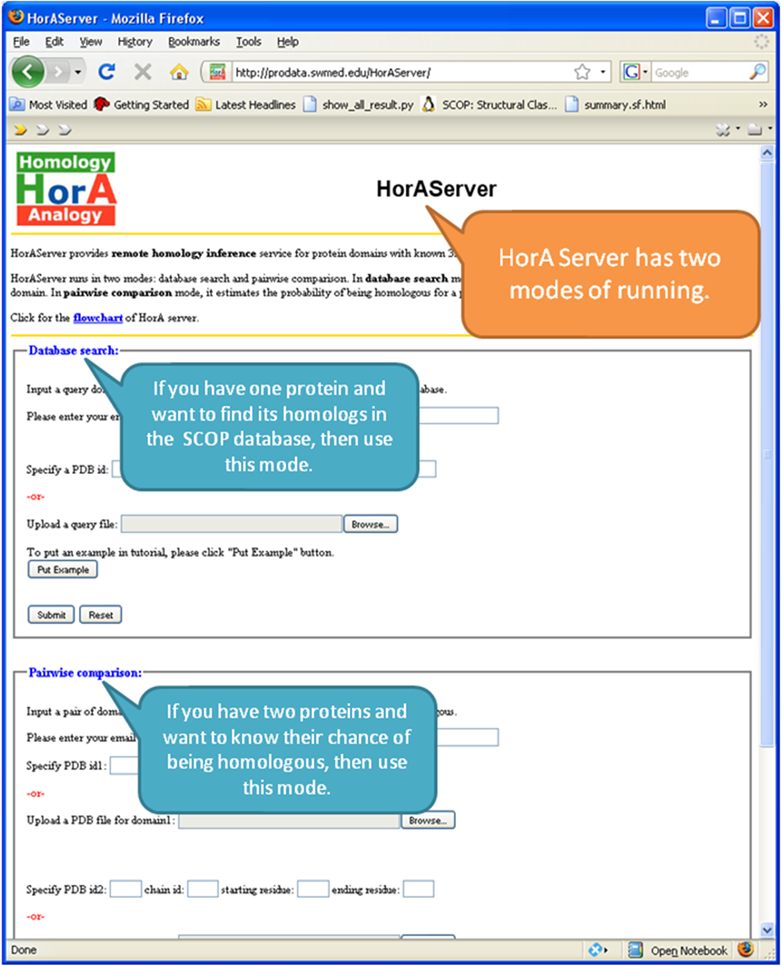
Database
Search
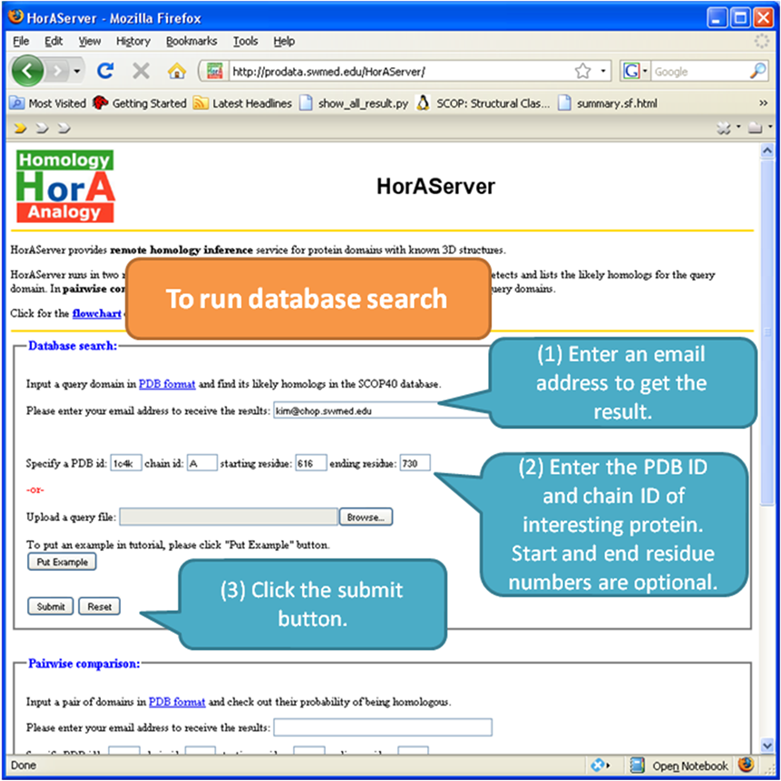
Database search mode compares the query protein (your protein of
interest) to proteins in SCOP
database (40% identity representatives), and you will get an email
containing the link to the result page like the one shown above.
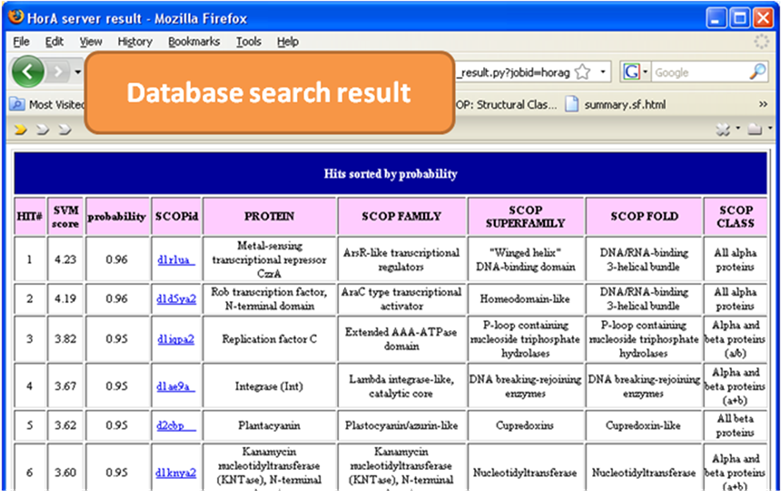
The result is a
table of proteins that are likely homologous to the query protein. More probable
homologs appear on the top and less probable homologs appear at the bottom. The
SVM score is a predictive score from
the model built to find homologous proteins combining many different sequence
similarity scores and structural similarity scores (details in reference). The probability conveniently measures how
probable the hit protein (from database) is homologous to the query protein. The
rest of the columns are information about the hit protein. SCOPid is the hit protein’s (or domain’s) unique identifier
in SCOP database. Protein, SCOP family, SCOP superfamily, SCOP fold,
and SCOP class show the
classification of the hit protein in the SCOP database.
Pairwise Comparison
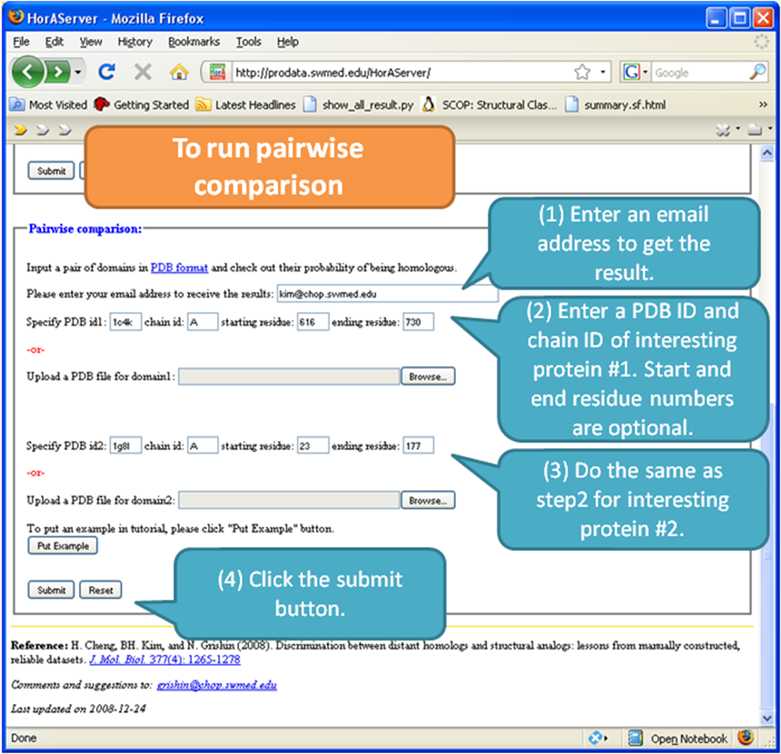
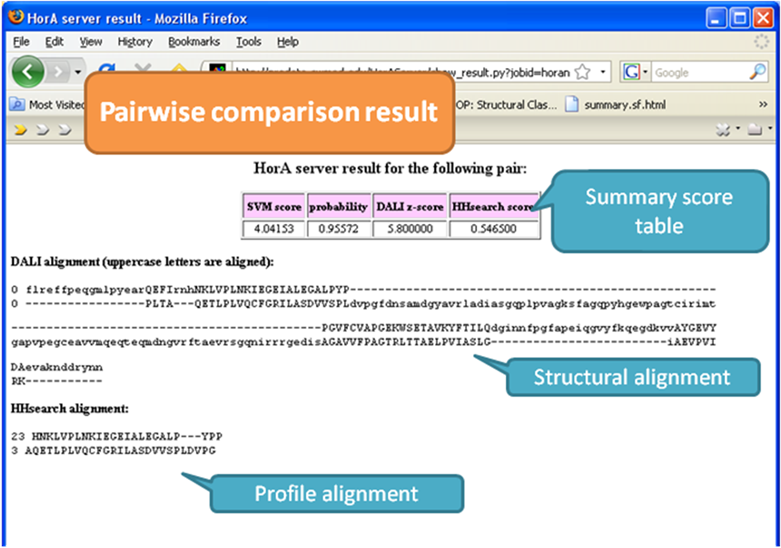
Pairwise
comparison mode compares two proteins submitted, and you will get an email
containing the link to the result page like shown above.
The result shows similarity scores between the two proteins of interest and
the alignments between them. The SVM
score is predictive score from the model built to find homologous proteins
combining many different sequence similarity scores and structural similarity
scores (details in reference). The probability
measures how probable the two proteins of interest are homologous. DALI Z-score is Z-score from DALI structural comparison
program developed by Holm
group. HHsearch score is
probability from HHsearch
HMM comparison program developed by Soding group.