Gonadotropin-regulated testicular RNA helicase
UniProt
Show FASTA Format
>gi|56749811|sp|Q9QY15.2|DDX25_MOUSE RecName: Full=ATP-dependent RNA helicase DDX25; AltName: Full=DEAD box protein 25; AltName: Full=Gonadotropin-regulated testicular RNA helicase
MASLLWGGDAGAAESERLNSHFSNLVHPRKNLRGIRSTTVPNIDGSLNTEDDDDDEDDVVDLAANSLLNK
LIRQSLIESSHRVEVLQKDPSSPLYSVKTFEELRLKEELLKGIYAMGFNRPSKIQEMALPMMLAHPPQNL
IAQSQSGTGKTAAFVLAMLSRVNALELFPQCLCLAPTYELALQTGRVVERMGKFCVDVEVMYAIRGNRIP
RGTEVTKQIIIGTPGTVLDWCFKRKLIDLTKIRVFVLDEADVMIDTQGFSDQSIRIQRALPSECQMLLFS
ATFEDSVWQFAERIIPDPNVIKLRKEELTLNNIRQYYVLCENRKGKYQALCNIYGGITIGQAIIFCQTRR
NAKWLTVEMMQDGHQVSLLSGELTVEQRASIIQRFRDGKEKVLITTNVCARGIDVKQVTIVVNFDLPVNQ
SEEPDYETYLHRIGRTGRFGKKGLAFNMIEVDKLPLLMKIQDHFNSNIKQLDPEDMDEIEKIEY
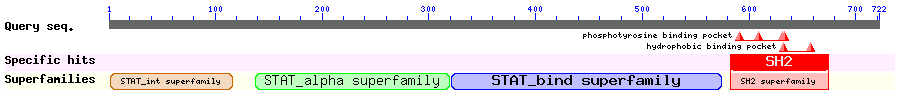
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 965545776001224531025787878876657888887678999999999999871011
Pred: CCCCCCCCCCCHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHH
AA: MASLLWGGDAGAAESERLNSHFSNLVHPRKNLRGIRSTTVPNIDGSLNTEDDDDDEDDVV
10 20 30 40 50 60
Conf: 056644666864201357772327860499998523233111599968884675239999
Pred: HHHHHHHHHHHHHCCCCCCCCEEEEEECCCCCCCCCCCCCCCCCCCHHHHHHHHHCCCCC
AA: DLAANSLLNKLIRQSLIESSHRVEVLQKDPSSPLYSVKTFEELRLKEELLKGIYAMGFNR
70 80 90 100 110 120
Conf: 903477320211269998621113567616799998864123499999808997436577
Pred: CCHHHHHHHCCCCCCCCCCCEEECCCCCCHHHHHHHHHHHCCCCCCCCCCEEEECCCHHH
AA: PSKIQEMALPMMLAHPPQNLIAQSQSGTGKTAAFVLAMLSRVNALELFPQCLCLAPTYEL
130 140 150 160 170 180
Conf: 877999999963455574688996299888897333218981582232311002333588
Pred: HHHHHHHHHHHCCCCCCCEEEEEECCCCCCCCCCCCCEEEECCCCHHHHHHHCCCCCCCC
AA: ALQTGRVVERMGKFCVDVEVMYAIRGNRIPRGTEVTKQIIIGTPGTVLDWCFKRKLIDLT
190 200 210 220 230 240
Conf: 315999623023330489984277997419978738998312763799999876599947
Pred: CCEEEEEECCHHHHHCCCCCCHHHHHHHHCCCCCCEEEEECCCCHHHHHHHHHHCCCCCE
AA: KIRVFVLDEADVMIDTQGFSDQSIRIQRALPSECQMLLFSATFEDSVWQFAERIIPDPNV
250 260 270 280 290 300
Conf: 876175357577435889748910169999974311131005898501332899999997
Pred: EEECCCCCCCCCCCEEEEECCCCCHHHHHHHHHHCCCCCEEEEEEECCCCCHHHHHHHHH
AA: IKLRKEELTLNNIRQYYVLCENRKGKYQALCNIYGGITIGQAIIFCQTRRNAKWLTVEMM
310 320 330 340 350 360
Conf: 579649984179897999999986308994058841432346543642499725889999
Pred: HCCCEEEEEECCCCHHHHHHHHHHHHCCCCEEEEECCCCCCCCCCCCEEEEEECCCCCCC
AA: QDGHQVSLLSGELTVEQRASIIQRFRDGKEKVLITTNVCARGIDVKQVTIVVNFDLPVNQ
370 380 390 400 410 420
Conf: 999995432213468898999613899432799888999999869984434999933331
Pred: CCCCCCCCCCCCCCCCCCCCCCCEEEEEEECCCHHHHHHHHHHHCCCCEECCCCCHHHHH
AA: SEEPDYETYLHRIGRTGRFGKKGLAFNMIEVDKLPLLMKIQDHFNSNIKQLDPEDMDEIE
430 440 450 460 470 480
Conf: 0269
Pred: CCCC
AA: KIEY
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 S -1.000 *
4 L -1.000 *
5 L -1.000 *
6 W -1.000 *
7 G -1.000 *
8 G -1.000 *
9 D -1.000 *
10 A -1.000 *
11 G -1.000 *
12 A -1.000 *
13 A -1.000 *
14 E -1.000 *
15 S -1.000 *
16 E -1.000 *
17 R -1.000 *
18 L -1.000 *
19 N -1.000 *
20 S -1.000 *
21 H -1.000 *
22 F -1.000 *
23 S -1.000 *
24 N -1.000 *
25 L -1.000 *
26 V -1.000 *
27 H -1.000 *
28 P -1.000 *
29 R -1.000 *
30 K -1.000 *
31 N -1.000 *
32 L -1.000 *
33 R -1.000 *
34 G -1.000 *
35 I -1.000 *
36 R -1.000 *
37 S -1.000 *
38 T -1.000 *
39 T -1.000 *
40 V -1.000 *
41 P -1.000 *
42 N -1.000 *
43 I -1.000 *
44 D -1.000 *
45 G -1.000 *
46 S -1.000 *
47 L -1.000 *
48 N -1.000 *
49 T -1.000 *
50 E -1.000 *
51 D -1.000 *
52 D -1.000 *
53 D -1.000 *
54 D -1.000 *
55 D -1.000 *
56 E -1.000 *
57 D -1.000 *
58 D -1.000 *
59 V -1.000 *
60 V -1.000 *
61 D -1.000 *
62 L -1.000 *
63 A -1.000 *
64 A -1.000 *
65 N -1.000 *
66 S -1.000 *
67 L -1.000 *
68 L -1.000 *
69 N -1.000 *
70 K -1.000 *
71 L -1.000 *
72 I -1.000 *
73 R -1.000 *
74 Q -1.000 *
75 S -1.000 *
76 L -1.000 *
77 I -1.000 *
78 E -1.000 *
79 S -1.106
80 S -0.900
81 H -1.206
82 R -0.904
83 V -0.789
84 E -0.688
85 V -0.904
86 L -0.857
87 Q -0.921
88 K -1.140
89 D -0.663
90 P -0.625
91 S -0.718
92 S -0.709
93 P -0.762
94 L -0.544
95 Y -0.985
96 S -0.493
97 V -1.036
98 K -0.984
99 T -0.516
100 F 2.417
101 E -0.665
102 E -0.510
103 L 0.847
104 R -0.831
105 L 0.801
106 K -1.078
107 E -0.613
108 E -0.837
109 L 0.622
110 L -0.590
111 K -0.274
112 G 0.869
113 I 0.844
114 Y -1.102
115 A -0.950
116 M -0.552
117 G 0.459
118 F 1.544
119 N -0.726
120 R -0.797
121 P 0.770
122 S 1.581
123 K -0.244
124 I 1.676
125 Q 2.417
126 E -0.351
127 M -0.761
128 A 0.183
129 L 0.929
130 P -0.638
131 M -1.031
132 M -0.464
133 L 0.008
134 A -0.959
135 H -0.665
136 P -1.047
137 P -1.000 *
138 Q -0.682
139 N 1.160
140 L 0.038
141 I 0.683
142 A 0.779
143 Q 0.931
144 S 1.522
145 Q 2.040
146 S 1.196
147 G 1.712
148 T 1.518
149 G 2.039
150 K 2.417
151 T 1.984
152 A 0.753
153 A 0.153
154 F 1.203
155 V -0.138
156 L 0.518
157 A 0.030
158 M -0.143
159 L 0.194
160 S -0.515
161 R -0.922
162 V 0.217
163 N -0.166
164 A -1.205
165 L -0.846
166 E -1.271
167 L -1.114
168 F -1.324
169 P 0.073
170 Q 1.395
171 C -0.295
172 L 0.607
173 C 0.087
174 L 0.750
175 A -0.001
176 P 1.329
177 T 0.607
178 Y 0.899
179 E 1.685
180 L 1.714
181 A 1.046
182 L -0.843
183 Q 2.040
184 T 0.221
185 G -0.922
186 R -0.703
187 V 0.279
188 V -0.261
189 E -1.194
190 R -0.944
191 M 0.216
192 G 0.076
193 K -0.873
194 F -0.579
195 C -1.039
196 V -1.026
197 D -0.687
198 V -0.135
199 E -0.989
200 V -0.636
201 M -1.210
202 Y -0.876
203 A -0.141
204 I -0.072
205 R -0.168
206 G -0.459
207 N -0.874
208 R -1.147
209 I -0.914
210 P -1.086
211 R -0.738
212 G -0.742
213 T -0.919
214 E -0.965
215 V -1.123
216 T -1.036
217 K -0.958
218 Q -0.256
219 I 0.595
220 I 0.793
221 I 0.495
222 G 1.744
223 T 1.713
224 P 1.840
225 G 2.417
226 T 0.055
227 V 0.138
228 L -0.861
229 D 0.701
230 W -0.045
231 C -0.363
232 F -1.074
233 K -0.322
234 R -0.740
235 K -0.239
236 L -0.938
237 I 0.425
238 D -0.921
239 L -0.509
240 T -0.618
241 K -1.062
242 I 0.292
243 R -0.858
244 V -0.655
245 F 0.073
246 V 0.585
247 L 1.003
248 D 2.417
249 E 2.417
250 A 1.433
251 D 2.417
252 V 0.003
253 M 1.669
254 I 0.654
255 D -0.323
256 T -0.768
257 Q -0.405
258 G 0.915
259 F 0.362
260 S -0.708
261 D -0.248
262 Q 0.333
263 S -0.012
264 I -0.950
265 R -0.927
266 I 0.445
267 Q -0.412
268 R -0.725
269 A -1.276
270 L -0.027
271 P -0.500
272 S -0.994
273 E -0.945
274 C -0.887
275 Q 1.279
276 M -0.394
277 L -0.204
278 L 0.644
279 F 0.346
280 S 2.417
281 A 2.042
282 T 2.417
283 F 0.116
284 E -0.254
285 D -1.012
286 S -1.061
287 V 0.842
288 W -0.997
289 Q -0.841
290 F 0.116
291 A 0.484
292 E -1.003
293 R -0.999
294 I 0.044
295 I -0.357
296 P -0.741
297 D -0.608
298 P 0.864
299 N -0.914
300 V -0.984
301 I 0.429
302 K -0.650
303 L 1.016
304 R -0.681
305 K -0.795
306 E -0.488
307 E -0.368
308 L -0.506
309 T 0.635
310 L -0.134
311 N -0.681
312 N -0.523
313 I 1.042
314 R -0.533
315 Q 1.935
316 Y -0.812
317 Y 0.108
318 V -0.327
319 L -1.138
320 C 0.036
321 E -1.081
322 N -0.794
323 R -1.179
324 K -0.632
325 G -1.139
326 K 1.590
327 Y -0.684
328 Q -0.631
329 A -0.418
330 L 0.495
331 C -0.501
332 N -0.412
333 I 0.423
334 Y 0.217
335 G -0.404
336 G -0.806
337 I -0.719
338 T -0.722
339 I -0.070
340 G -0.537
341 Q 0.082
342 A -0.201
343 I 0.569
344 I 0.700
345 F 1.513
346 C -0.041
347 Q -0.451
348 T 0.362
349 R 0.744
350 R -0.943
351 N -0.954
352 A 0.590
353 K -0.922
354 W -1.117
355 L 0.905
356 T -0.891
357 V -1.087
358 E -0.889
359 M 1.521
360 M -1.051
361 Q -0.912
362 D -0.670
363 G 0.197
364 H 0.339
365 Q -0.980
366 V 0.228
367 S -0.570
368 L -0.826
369 L 0.618
370 S 0.553
371 G 1.303
372 E -0.164
373 L -0.448
374 T -0.938
375 V -0.608
376 E -1.031
377 Q -0.754
378 R 2.417
379 A -0.354
380 S -0.999
381 I 0.205
382 I 0.372
383 Q -0.653
384 R -0.642
385 F 0.847
386 R 1.109
387 D -0.924
388 G 0.438
389 K -0.985
390 E -0.780
391 K 0.271
392 V 0.538
393 L 1.543
394 I 1.201
395 T 0.675
396 T 2.040
397 N 1.522
398 V 1.119
399 C -0.275
400 A 0.998
401 R 2.039
402 G 2.035
403 I 1.306
404 D 2.035
405 V 1.536
406 K -0.830
407 Q -0.502
408 V 1.026
409 T 0.083
410 I -0.182
411 V 1.664
412 V 0.220
413 N 1.592
414 F 1.078
415 D 0.294
416 L 0.380
417 P 2.034
418 V -1.110
419 N -0.903
420 Q -1.141
421 S -1.000 *
422 E -1.000 *
423 E -1.000 *
424 P -0.266
425 D 0.778
426 Y -1.139
427 E 0.603
428 T -0.297
429 Y 2.035
430 L 0.655
431 H 2.417
432 R 2.417
433 I 1.751
434 G 2.035
435 R 2.035
436 T 0.692
437 G 1.892
438 R 2.417
439 F 0.866
440 G 1.412
441 K 0.071
442 K -0.968
443 G 2.035
444 L -0.978
445 A 1.235
446 F 0.052
447 N 0.428
448 M 0.706
449 I 0.490
450 E -0.997
451 V -1.029
452 D -0.884
453 K -0.498
454 L -1.052
455 P -1.178
456 L -1.241
457 L 0.205
458 M -0.961
459 K -1.120
460 I 0.604
461 Q -0.215
462 D -0.864
463 H -0.722
464 F -0.223
465 N -0.600
466 S -0.766
467 N -1.044
468 I 0.388
469 K -1.073
470 Q -1.059
471 L -0.791
472 D -0.898
473 P -0.645
474 E -0.824
475 D -0.528
476 M -1.113
477 D -0.587
478 E -0.721
479 I 0.235
480 E -0.171
481 K -0.339
482 I -1.000 *
483 E -1.000 *
484 Y -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 102.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MASLLWGGDA GAAESERLNS HFSNLVHPRK NLRGIRSTTV PNIDGSLNTE DDDDDEDDVV
70 80 90 100 110 120
DLAANSLLNK LIRQSLIESS HRVEVLQKDP SSPLYSVKTF EELRLKEELL KGIYAMGFNR
130 140 150 160 170 180
PSKIQEMALP MMLAHPPQNL IAQSQSGTGK TAAFVLAMLS RVNALELFPQ CLCLAPTYEL
190 200 210 220 230 240
ALQTGRVVER MGKFCVDVEV MYAIRGNRIP RGTEVTKQII IGTPGTVLDW CFKRKLIDLT
250 260 270 280 290 300
KIRVFVLDEA DVMIDTQGFS DQSIRIQRAL PSECQMLLFS ATFEDSVWQF AERIIPDPNV
310 320 330 340 350 360
IKLRKEELTL NNIRQYYVLC ENRKGKYQAL CNIYGGITIG QAIIFCQTRR NAKWLTVEMM
370 380 390 400 410 420
QDGHQVSLLS GELTVEQRAS IIQRFRDGKE KVLITTNVCA RGIDVKQVTI VVNFDLPVNQ
430 440 450 460 470 480
SEEPDYETYL HRIGRTGRFG KKGLAFNMIE VDKLPLLMKI QDHFNSNIKQ LDPEDMDEIE
KIEY
[1]. "Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis."
Sheng Y, Tsai-Morris CH, Gutti R, Maeda Y, Dufau ML. (2006)
J Biol Chem,
281:35048-56
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
