Bcl-2-related ovarian killer protein
UniProt
Show FASTA Format
>gi|57012554|sp|Q9UMX3.1|BOK_HUMAN RecName: Full=Bcl-2-related ovarian killer protein; Short=hBOK; AltName: Full=Bcl-2-like protein 9; Short=Bcl2-L-9
MEVLRRSSVFAAEIMDAFDRSPTDKELVAQAKALGREYVHARLLRAGLSWSAPERAAPVPGRLAEVCAVL
LRLGDELEMIRPSVYRNVARQLHISLQSEPVVTDAFLAVAGHIFSAGITWGKVVSLYAVAAGLAVDCVRQ
AQPAMVHALVDCLGEFVRKTLATWLRRRGGWTDVLKCVVSTDPGLRSHWLVAALCSFGRFLKAAFFVLLP
ER
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 901121015889752029999996999999999998889978722689999886679999
Pred: CHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCC
AA: MEVLRRSSVFAAEIMDAFDRSPTDKELVAQAKALGREYVHARLLRAGLSWSAPERAAPVP
10 20 30 40 50 60
Conf: 743566489978899998448456687899743379995008999999998751079983
Pred: CCHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHCCCCCH
AA: GRLAEVCAVLLRLGDELEMIRPSVYRNVARQLHISLQSEPVVTDAFLAVAGHIFSAGITW
70 80 90 100 110 120
Conf: 789999998779999999833835698999999999884799999750788888764405
Pred: HHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHCC
AA: GKVVSLYAVAAGLAVDCVRQAQPAMVHALVDCLGEFVRKTLATWLRRRGGWTDVLKCVVS
130 140 150 160 170 180
Conf: 89787326889999999789987765425579
Pred: CCCCCCHHHHHHHHHHHHHHHHHHHHHHCCCC
AA: TDPGLRSHWLVAALCSFGRFLKAAFFVLLPER
190 200 210
Show Conservation Score by AL2CO
1 M -1.000 *
2 E -1.000 *
3 V -1.000 *
4 L -1.000 *
5 R -1.000 *
6 R -1.000 *
7 S -1.000 *
8 S -1.000 *
9 V -1.000 *
10 F -1.000 *
11 A -1.000 *
12 A -1.000 *
13 E -1.000 *
14 I -1.000 *
15 M -1.000 *
16 D -1.000 *
17 A -1.000 *
18 F -1.000 *
19 D -1.000 *
20 R -1.000 *
21 S -1.000 *
22 P -1.000 *
23 T -1.000 *
24 D -1.000 *
25 K -1.000 *
26 E -1.000 *
27 L 0.380
28 V -0.161
29 A -0.761
30 Q 0.410
31 A 1.225
32 K -0.637
33 A -0.819
34 L 1.351
35 G 0.009
36 R -0.777
37 E -0.191
38 Y 1.564
39 V 0.538
40 H -0.779
41 A -0.667
42 R -0.332
43 L 0.115
44 L -0.514
45 R -0.168
46 A -0.701
47 G 0.083
48 L -0.805
49 S -0.872
50 W -1.033
51 S -0.676
52 A -0.787
53 P -0.753
54 E -0.616
55 R -0.935
56 A -1.072
57 A -0.781
58 P -0.557
59 V -0.571
60 P -0.815
61 G -0.563
62 R -0.605
63 L -0.920
64 A -0.816
65 E -0.625
66 V 0.040
67 C -0.951
68 A -0.792
69 V -0.916
70 L 0.216
71 L -0.587
72 R -0.948
73 L 0.572
74 G 0.992
75 D -0.516
76 E -0.506
77 L 1.988
78 E -0.037
79 M -0.630
80 I -0.413
81 R -0.022
82 P -0.496
83 S -0.801
84 V -0.488
85 Y 0.135
86 R -0.260
87 N -0.124
88 V 0.004
89 A -0.492
90 R -0.673
91 Q -0.319
92 L -0.198
93 H -0.957
94 I -0.299
95 S -0.330
96 L -1.078
97 Q -0.962
98 S -0.563
99 E -0.734
100 P -0.839
101 V -0.579
102 V 0.265
103 T -0.811
104 D -0.452
105 A -0.649
106 F 1.442
107 L -0.843
108 A -0.407
109 V 1.203
110 A 0.336
111 G -0.605
112 H -0.160
113 I 1.104
114 F 1.014
115 S -0.390
116 A -0.239
117 G 0.625
118 I 0.696
119 T 1.449
120 W 2.808
121 G 1.691
122 K 0.948
123 V 2.506
124 V 1.883
125 S 1.238
126 L 1.585
127 Y 1.326
128 A -0.021
129 V 1.770
130 A 1.218
131 A 0.666
132 G -0.719
133 L 1.735
134 A 1.159
135 V -0.359
136 D -0.097
137 C -0.269
138 V -0.477
139 R -0.181
140 Q -0.406
141 A -0.186
142 Q -0.721
143 P -0.882
144 A -0.492
145 M -0.895
146 V 0.205
147 H -0.791
148 A -0.946
149 L 2.258
150 V -0.183
151 D -0.547
152 C -0.424
153 L 0.793
154 G -0.219
155 E -0.299
156 F 1.195
157 V 1.468
158 R -0.632
159 K -0.349
160 T -0.601
161 L 0.408
162 A -0.862
163 T -0.670
164 W 3.634
165 L 2.216
166 R -0.469
167 R -0.155
168 R 0.779
169 G 2.045
170 G 3.182
171 W 5.103
172 T -0.450
173 D -0.071
174 V 1.013
175 L -0.619
176 K -0.337
177 C -0.819
178 V -0.277
179 V -0.948
180 S -0.340
181 T -0.873
182 D -0.397
183 P -0.552
184 G -0.782
185 L -0.935
186 R -0.902
187 S -0.876
188 H -1.017
189 W 0.153
190 L -0.442
191 V -0.161
192 A -0.307
193 A 0.295
194 L 0.336
195 C -0.158
196 S 0.037
197 F 0.394
198 G 0.107
199 R -0.465
200 F 0.579
201 L 0.769
202 K 0.695
203 A -1.000 *
204 A -1.000 *
205 F -1.000 *
206 F -1.000 *
207 V -1.000 *
208 L -1.000 *
209 L -1.000 *
210 P -1.000 *
211 E -1.000 *
212 R -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 107.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MEVLRRSSVF AAEIMDAFDR SPTDKELVAQ AKALGREYVH ARLLRAGLSW SAPERAAPVP
70 80 90 100 110 120
GRLAEVCAVL LRLGDELEMI RPSVYRNVAR QLHISLQSEP VVTDAFLAVA GHIFSAGITW
130 140 150 160 170 180
GKVVSLYAVA AGLAVDCVRQ AQPAMVHALV DCLGEFVRKT LATWLRRRGG WTDVLKCVVS
190 200 210
TDPGLRSHWL VAALCSFGRF LKAAFFVLLP ER
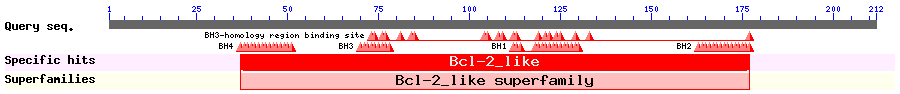
Bok is a pro-apoptotic member of the Bcl-2 family of proteins that is the key regulator of the intrinsic apoptotic pathway. It contains Bcl-2 Homology (BH) domains 1-3 and a C-terminal transmembrane domain. Bartholomeusz et al. showed that in the BH3 domain of Bok, there is a NES. LMB treatment or point mutations of the NES increased the nuclear accumulation and pro-apoptotic potency of Bok.
[1]. "Nuclear translocation of the pro-apoptotic Bcl-2 family member Bok induces apoptosis."
Bartholomeusz G, Wu Y, Ali Seyed M, Xia W, Kwong KY, Hortobagyi G, Hung MC (2006)
Mol Carcinog,
45:73-83
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
