T-box transcription factor TBX5
UniProt
Unknown (the 3D structure listed below is for Tbx3 and the secondary structure of the NES region was not determined)
Show FASTA Format
>gi|82175283|sp|Q9PWE8.1|TBX5_CHICK RecName: Full=T-box transcription factor TBX5; Short=T-box protein 5
MADTEEGFGLPSTPVDSEAKELQAEAKQDPQLGTTSKAPTSPQAAFTQQGMEGIKVFLHERELWLKFHEV
GTEMIITKAGRRMFPSYKVKVTGLNPKTKYILLMDIVPADDHRYKFADNKWSVTGKAEPAMPGRLYVHPD
SPATGAHWMRQLVSFQKLKLTNNHLDPFGHIILNSMHKYQPRLHIVKADENNGFGSKNTAFCTHVFPETA
FIAVTSYQNHKITQLKIENNPFAKGFRGSDDMELHRMSRMQSKEYPVVPRSTVRQKVSSNHSPFSGETRV
LSTSSNLGSQYQCENGVSSTSQDLLPPTNPYPISQEHSQIYHCTKRKDEECSTTEHPYKKPYMETSPAEE
DPFYRSSYPQQQGLNTSYRTESAQRQACMYASSAPPTDPVPSLEDISCNTWPSVPSYSSCTVSAMQPMDR
LPYQHFSAHFTSGPLMPRLSSVANHTSPQIGDTHSMFQHQTSVSHQPIVRQCGPQTGIQSPPSSLQPAEF
LYSHGVPRTLSPHQYHSVHGVGMVPEWSENS
Show Domain Info by CDD
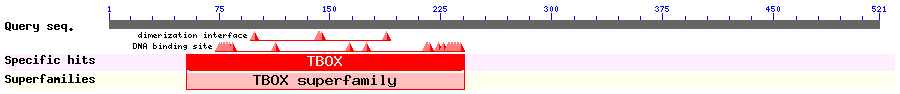
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 987677899999999940332102458999999999999999851112564621798620
Pred: CCCCCCCCCCCCCCCCCHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCEEEEEC
AA: MADTEEGFGLPSTPVDSEAKELQAEAKQDPQLGTTSKAPTSPQAAFTQQGMEGIKVFLHE
10 20 30 40 50 60
Conf: 478999974075078923695247650399707795762789985223499742115881
Pred: HHHHHHHHHCCCEEEEECCCCCCCCCCEEEEECCCCCCCEEEEEEEEECCCCCCCCCCCC
AA: RELWLKFHEVGTEMIITKAGRRMFPSYKVKVTGLNPKTKYILLMDIVPADDHRYKFADNK
70 80 90 100 110 120
Conf: 245024899999941559999871666521013200010114677988845516876788
Pred: EEEECCCCCCCCCCEEECCCCCCCHHHHHHCCCCCCCEECCCCCCCCCCCEEECCCCCCC
AA: WSVTGKAEPAMPGRLYVHPDSPATGAHWMRQLVSFQKLKLTNNHLDPFGHIILNSMHKYQ
130 140 150 160 170 180
Conf: 872089647888889877675123212541267521468500000013684456766899
Pred: CCCCEEECCCCCCCCCCCCCCCEEEECCCCEEEEECCCCCCCCCCCCCCCCCCCCCCCCC
AA: PRLHIVKADENNGFGSKNTAFCTHVFPETAFIAVTSYQNHKITQLKIENNPFAKGFRGSD
190 200 210 220 230 240
Conf: 703444200246889999974212578899999999887678998889977566999888
Pred: CCHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: DMELHRMSRMQSKEYPVVPRSTVRQKVSSNHSPFSGETRVLSTSSNLGSQYQCENGVSST
250 260 270 280 290 300
Conf: 889899999999998884333456567889999999998999999999989998899998
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: SQDLLPPTNPYPISQEHSQIYHCTKRKDEECSTTEHPYKKPYMETSPAEEDPFYRSSYPQ
310 320 330 340 350 360
Conf: 899999998975100147778999999999999865579999878999897677799988
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: QQGLNTSYRTESAQRQACMYASSAPPTDPVPSLEDISCNTWPSVPSYSSCTVSAMQPMDR
370 380 390 400 410 420
Conf: 999788888989999998422455899999998877767888886763235899999899
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: LPYQHFSAHFTSGPLMPRLSSVANHTSPQIGDTHSMFQHQTSVSHQPIVRQCGPQTGIQS
430 440 450 460 470 480
Conf: 99999997541247899998998764125678768797899
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: PPSSLQPAEFLYSHGVPRTLSPHQYHSVHGVGMVPEWSENS
490 500 510 520
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 D -1.000 *
4 T -1.000 *
5 E -1.000 *
6 E -1.000 *
7 G -1.000 *
8 F -1.000 *
9 G -1.000 *
10 L -1.000 *
11 P -1.000 *
12 S -1.000 *
13 T -1.000 *
14 P -1.000 *
15 V -1.000 *
16 D -1.000 *
17 S -1.000 *
18 E -0.641
19 A -0.681
20 K -0.707
21 E -0.554
22 L -0.776
23 Q -0.537
24 A -0.696
25 E -0.630
26 A -0.454
27 K -0.592
28 Q -0.880
29 D -0.675
30 P -0.712
31 Q -0.625
32 L -0.580
33 G -0.604
34 T -0.651
35 T -0.767
36 S -0.850
37 K -0.744
38 A -0.693
39 P -0.471
40 T -0.547
41 S -0.774
42 P -0.694
43 Q -0.736
44 A -1.000 *
45 A -1.000 *
46 F -1.000 *
47 T -1.000 *
48 Q -1.000 *
49 Q -1.000 *
50 G -1.000 *
51 M -0.846
52 E -0.493
53 G -0.396
54 I 0.340
55 K -0.353
56 V 0.439
57 F -0.732
58 L 2.184
59 H 0.199
60 E -0.169
61 R -0.658
62 E -0.234
63 L 1.717
64 W 2.245
65 L -0.632
66 K 0.028
67 F 2.245
68 H -0.173
69 E -0.189
70 V -0.481
71 G 0.371
72 T 1.644
73 E 1.886
74 M 2.200
75 I 1.138
76 I 1.175
77 T 1.887
78 K 1.893
79 A 0.228
80 G 2.252
81 R 1.781
82 R 1.908
83 M 1.225
84 F 1.610
85 P 2.267
86 S -0.315
87 Y 0.210
88 K 0.711
89 V 0.093
90 K -0.041
91 V 0.429
92 T -0.487
93 G 1.438
94 L 0.605
95 N -0.050
96 P -0.126
97 K -0.781
98 T -0.219
99 K -0.031
100 Y 1.903
101 I -0.295
102 L 0.621
103 L 0.473
104 M 0.639
105 D 1.945
106 I 0.725
107 V 0.115
108 P 0.121
109 A -0.229
110 D 1.274
111 D 0.041
112 H -0.217
113 R 0.907
114 Y 0.889
115 K 1.341
116 F 1.131
117 A -0.261
118 D -0.111
119 N -0.129
120 K -0.538
121 W 2.132
122 S -0.020
123 V 0.028
124 T 0.287
125 G 1.877
126 K 0.316
127 A 0.523
128 E 1.829
129 P 0.222
130 A -0.464
131 M -0.207
132 P 0.108
133 G -0.521
134 R 0.372
135 L -0.169
136 Y 1.011
137 V 0.183
138 H 1.879
139 P 1.882
140 D 1.337
141 S 1.885
142 P 1.218
143 A 0.641
144 T -0.374
145 G 2.677
146 A -0.270
147 H -0.005
148 W 1.399
149 M -0.037
150 R 0.322
151 Q -0.179
152 L -0.172
153 V 0.662
154 S 0.311
155 F 1.891
156 Q -0.273
157 K 1.671
158 L 0.802
159 K 2.677
160 L 0.876
161 T 1.149
162 N 1.040
163 N 1.041
164 H -0.568
165 L 0.101
166 D 0.651
167 P -0.278
168 F -0.475
169 G 0.251
170 H -0.111
171 I 0.371
172 I 0.914
173 L 1.518
174 N 0.584
175 S 1.726
176 M 1.609
177 H 1.838
178 K 1.828
179 Y 1.845
180 Q 0.029
181 P 1.837
182 R 1.697
183 L 0.503
184 H 1.843
185 I 0.958
186 V 0.840
187 K -0.241
188 A -0.384
189 D -0.296
190 E -0.235
191 N -0.623
192 N -0.662
193 G -0.562
194 F -0.587
195 G -0.492
196 S -0.695
197 K -0.716
198 N -0.553
199 T -0.447
200 A -0.722
201 F -0.240
202 C -0.551
203 T 0.323
204 H 0.073
205 V -0.356
206 F 2.225
207 P -0.182
208 E 0.446
209 T 1.490
210 A -0.419
211 F 1.845
212 I 0.250
213 A 1.013
214 V 2.240
215 T 2.677
216 S 1.613
217 Y 2.220
218 Q 2.220
219 N 1.691
220 H -0.382
221 K 0.017
222 I 1.137
223 T 0.855
224 Q 0.446
225 L 2.677
226 K 2.677
227 I 1.506
228 E 0.357
229 N -0.000
230 N 2.677
231 P 2.677
232 F 2.677
233 A 1.531
234 K 1.199
235 G 1.441
236 F 1.531
237 R 0.801
238 G 0.422
239 S -0.636
240 D -0.137
241 D -0.434
242 M -0.570
243 E -0.667
244 L -0.824
245 H -0.702
246 R -0.919
247 M -0.830
248 S -0.841
249 R -0.819
250 M -0.797
251 Q -0.717
252 S -0.644
253 K -0.814
254 E -0.587
255 Y -0.891
256 P -0.809
257 V -0.774
258 V -0.790
259 P -0.880
260 R -0.727
261 S -0.835
262 T -0.822
263 V -0.833
264 R -0.917
265 Q -0.814
266 K -0.891
267 V -0.830
268 S -0.766
269 S -0.779
270 N -0.590
271 H -0.868
272 S -0.820
273 P -0.774
274 F -0.918
275 S -0.912
276 G -0.772
277 E -0.732
278 T -0.719
279 R -0.809
280 V -0.793
281 L -0.749
282 S -0.790
283 T -0.783
284 S -0.890
285 S -0.830
286 N -0.765
287 L -0.914
288 G -0.814
289 S -0.802
290 Q -0.828
291 Y -0.842
292 Q -0.684
293 C -0.844
294 E -0.624
295 N -0.748
296 G -0.878
297 V -0.793
298 S -0.836
299 S -0.826
300 T -0.864
301 S -0.660
302 Q -0.618
303 D -0.730
304 L -0.904
305 L -0.796
306 P -0.744
307 P -0.865
308 T -0.801
309 N -0.827
310 P -0.776
311 Y -0.857
312 P -0.766
313 I -0.813
314 S -0.823
315 Q -0.565
316 E -0.720
317 H -0.693
318 S -0.621
319 Q -0.696
320 I -0.818
321 Y -0.920
322 H -0.857
323 C -0.777
324 T -0.701
325 K -0.709
326 R -0.680
327 K -0.691
328 D -0.769
329 E -0.735
330 E -0.691
331 C -0.793
332 S -0.664
333 T -0.747
334 T -0.594
335 E -0.497
336 H -0.776
337 P -0.594
338 Y -0.841
339 K -0.646
340 K -0.826
341 P -0.641
342 Y -0.896
343 M -0.850
344 E -0.576
345 T -0.756
346 S -0.698
347 P -0.630
348 A -0.554
349 E -0.798
350 E -0.745
351 D -0.519
352 P -0.525
353 F -0.729
354 Y -0.775
355 R -0.891
356 S -0.729
357 S -0.588
358 Y -0.841
359 P -0.653
360 Q -0.835
361 Q -0.831
362 Q -0.769
363 G -0.533
364 L -0.554
365 N -0.688
366 T -0.476
367 S -0.789
368 Y -0.750
369 R -0.742
370 T -0.691
371 E -0.725
372 S -0.752
373 A -0.749
374 Q -0.647
375 R -1.000 *
376 Q -0.494
377 A -0.757
378 C -0.717
379 M -0.741
380 Y -0.675
381 A -0.491
382 S -0.773
383 S -0.619
384 A -0.609
385 P -0.624
386 P -0.564
387 T -1.000 *
388 D -1.000 *
389 P -1.000 *
390 V -1.000 *
391 P -1.000 *
392 S -1.000 *
393 L -1.000 *
394 E -1.000 *
395 D -1.000 *
396 I -1.000 *
397 S -1.000 *
398 C -1.000 *
399 N -1.000 *
400 T -1.000 *
401 W -1.000 *
402 P -1.000 *
403 S -1.000 *
404 V -1.000 *
405 P -1.000 *
406 S -1.000 *
407 Y -1.000 *
408 S -1.000 *
409 S -1.000 *
410 C -1.000 *
411 T -1.000 *
412 V -1.000 *
413 S -1.000 *
414 A -1.000 *
415 M -1.000 *
416 Q -1.000 *
417 P -1.000 *
418 M -1.000 *
419 D -1.000 *
420 R -1.000 *
421 L -1.000 *
422 P -1.000 *
423 Y -1.000 *
424 Q -1.000 *
425 H -1.000 *
426 F -1.000 *
427 S -1.000 *
428 A -1.000 *
429 H -1.000 *
430 F -1.000 *
431 T -1.000 *
432 S -1.000 *
433 G -1.000 *
434 P -1.000 *
435 L -1.000 *
436 M -1.000 *
437 P -1.000 *
438 R -1.000 *
439 L -1.000 *
440 S -1.000 *
441 S -1.000 *
442 V -1.000 *
443 A -1.000 *
444 N -1.000 *
445 H -1.000 *
446 T -1.000 *
447 S -1.000 *
448 P -1.000 *
449 Q -1.000 *
450 I -1.000 *
451 G -1.000 *
452 D -1.000 *
453 T -1.000 *
454 H -1.000 *
455 S -1.000 *
456 M -1.000 *
457 F -1.000 *
458 Q -1.000 *
459 H -1.000 *
460 Q -1.000 *
461 T -1.000 *
462 S -1.000 *
463 V -1.000 *
464 S -1.000 *
465 H -1.000 *
466 Q -1.000 *
467 P -1.000 *
468 I -1.000 *
469 V -1.000 *
470 R -1.000 *
471 Q -1.000 *
472 C -1.000 *
473 G -1.000 *
474 P -1.000 *
475 Q -1.000 *
476 T -1.000 *
477 G -1.000 *
478 I -1.000 *
479 Q -1.000 *
480 S -1.000 *
481 P -1.000 *
482 P -1.000 *
483 S -1.000 *
484 S -1.000 *
485 L -1.000 *
486 Q -1.000 *
487 P -1.000 *
488 A -1.000 *
489 E -1.000 *
490 F -1.000 *
491 L -1.000 *
492 Y -1.000 *
493 S -1.000 *
494 H -1.000 *
495 G -1.000 *
496 V -1.000 *
497 P -1.000 *
498 R -1.000 *
499 T -1.000 *
500 L -1.000 *
501 S -1.000 *
502 P -1.000 *
503 H -1.000 *
504 Q -1.000 *
505 Y -1.000 *
506 H -1.000 *
507 S -1.000 *
508 V -1.000 *
509 H -1.000 *
510 G -1.000 *
511 V -1.000 *
512 G -1.000 *
513 M -1.000 *
514 V -1.000 *
515 P -1.000 *
516 E -1.000 *
517 W -1.000 *
518 S -1.000 *
519 E -1.000 *
520 N -1.000 *
521 S -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 127.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MADTEEGFGL PSTPVDSEAK ELQAEAKQDP QLGTTSKAPT SPQAAFTQQG MEGIKVFLHE
70 80 90 100 110 120
RELWLKFHEV GTEMIITKAG RRMFPSYKVK VTGLNPKTKY ILLMDIVPAD DHRYKFADNK
130 140 150 160 170 180
WSVTGKAEPA MPGRLYVHPD SPATGAHWMR QLVSFQKLKL TNNHLDPFGH IILNSMHKYQ
190 200 210 220 230 240
PRLHIVKADE NNGFGSKNTA FCTHVFPETA FIAVTSYQNH KITQLKIENN PFAKGFRGSD
250 260 270 280 290 300
DMELHRMSRM QSKEYPVVPR STVRQKVSSN HSPFSGETRV LSTSSNLGSQ YQCENGVSST
310 320 330 340 350 360
SQDLLPPTNP YPISQEHSQI YHCTKRKDEE CSTTEHPYKK PYMETSPAEE DPFYRSSYPQ
370 380 390 400 410 420
QQGLNTSYRT ESAQRQACMY ASSAPPTDPV PSLEDISCNT WPSVPSYSSC TVSAMQPMDR
430 440 450 460 470 480
LPYQHFSAHF TSGPLMPRLS SVANHTSPQI GDTHSMFQHQ TSVSHQPIVR QCGPQTGIQS
490 500 510 520
PPSSLQPAEF LYSHGVPRTL SPHQYHSVHG VGMVPEWSEN S
2X6U (X-Ray,1.90 Å resolution)
Tbx5 protein is a transcription factor, which regulates the genes responsible for heart and limb development. Tbx5 has two NLSs and one NES. Both NLSs are required for efficient nuclear localization. During heart and limb development, Tbx5 is dynamically expressed in nuclear and/or cytoplasmic compartments. The interaction and association of Tbx5 with LMP4 can lead to Tbx5 localization along the actin cytoskeleton. Tbx5 can be co-immunoprecipitated with CRM1. The proposed NES region is conserved among all T-box family of proteins. Single mutation of the hydrophobic residues doesn't affect interactions with CRM1, but can prevent cytoplasmic localization in the presence of LMP4. LMP4 is exclusively cytoplasmic.
[1]. "An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor."
Kulisz A, Simon HG (2008)
Mol Cell Biol,
28:1553-64
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
