Calcium/calmodulin-dependent protein kinase type 1
UniProt
Show FASTA Format
>gi|3122310|sp|Q63450.2|KCC1A_RAT RecName: Full=Calcium/calmodulin-dependent protein kinase type 1; AltName: Full=CaM kinase I; Short=CaM-KI; AltName: Full=CaM kinase I alpha; Short=CaMKI-alpha
MPGAVEGPRWKQAEDIRDIYDFRDVLGTGAFSEVILAEDKRTQKLVAIKCIAKKALEGKEGSMENEIAVL
HKIKHPNIVALDDIYESGGHLYLIMQLVSGGELFDRIVEKGFYTERDASRLIFQVLDAVKYLHDLGIVHR
DLKPENLLYYSLDEDSKIMISDFGLSKMEDPGSVLSTACGTPGYVAPEVLAQKPYSKAVDCWSIGVIAYI
LLCGYPPFYDENDAKLFEQILKAEYEFDSPYWDDISDSAKDFIRHLMEKDPEKRFTCEQALQHPWIAGDT
ALDKNIHQSVSEQIKKNFAKSKWKQAFNATAVVRHMRKLQLGTSQEGQGQTASHGELLTPTAGGPAAGCC
CRDCCVEPGSELPPAPPPSSRAMD
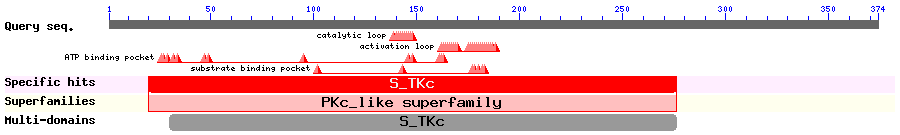
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 999999998776457300110100006776037999576168976998852204334632
Pred: CCCCCCCCCCCCCCCCCCHHHHHHHCCCCCCEEEEEEEECCCCCEEEEEEEEECCCCCCC
AA: MPGAVEGPRWKQAEDIRDIYDFRDVLGTGAFSEVILAEDKRTQKLVAIKCIAKKALEGKE
10 20 30 40 50 60
Conf: 014668999884389860001243202875888785112540356766315666354778
Pred: CHHHHHHHHHHHCCCCCEEEEEEEEECCCEEEEEEEECCCCCHHHHHHHCCCCCHHHHHH
AA: GSMENEIAVLHKIKHPNIVALDDIYESGGHLYLIMQLVSGGELFDRIVEKGFYTERDASR
70 80 90 100 110 120
Conf: 999999899865746974568998640156789999826545653322478983224568
Pred: HHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCEEECCCCCCCCCCCCCCCCCCCC
AA: LIFQVLDAVKYLHDLGIVHRDLKPENLLYYSLDEDSKIMISDFGLSKMEDPGSVLSTACG
130 140 150 160 170 180
Conf: 876522445215898874311344676775641999975686089999988152220389
Pred: CCCCCHHHHHCCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHCCCCCCCCC
AA: TPGYVAPEVLAQKPYSKAVDCWSIGVIAYILLCGYPPFYDENDAKLFEQILKAEYEFDSP
190 200 210 220 230 240
Conf: 861104318999997622298965678898419851378545677335699999986578
Pred: CCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHHH
AA: YWDDISDSAKDFIRHLMEKDPEKRFTCEQALQHPWIAGDTALDKNIHQSVSEQIKKNFAK
250 260 270 280 290 300
Conf: 999999889999999863004999889899778998789999999998777899888999
Pred: HHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: SKWKQAFNATAVVRHMRKLQLGTSQEGQGQTASHGELLTPTAGGPAAGCCCRDCCVEPGS
310 320 330 340 350 360
Conf: 99999999998999
Pred: CCCCCCCCCCCCCC
AA: ELPPAPPPSSRAMD
370
Show Conservation Score by AL2CO
1 M -1.000 *
2 P -1.000 *
3 G -1.000 *
4 A -1.000 *
5 V -1.000 *
6 E -1.000 *
7 G -1.000 *
8 P -1.000 *
9 R -1.000 *
10 W -1.000 *
11 K -0.811
12 Q -0.765
13 A -0.937
14 E -1.009
15 D -0.822
16 I 0.164
17 R -0.875
18 D -0.773
19 I -1.002
20 Y 1.174
21 D -0.904
22 F -0.115
23 R -0.827
24 D -0.477
25 V -0.231
26 L 1.031
27 G 1.729
28 T -0.494
29 G 2.059
30 A 0.014
31 F 0.793
32 S 0.903
33 E 0.136
34 V 1.204
35 I -0.326
36 L -0.662
37 A 0.530
38 E -0.610
39 D -0.550
40 K -0.174
41 R -0.945
42 T -0.215
43 Q -0.498
44 K -0.990
45 L -1.091
46 V -0.346
47 A 2.060
48 I 0.202
49 K 2.440
50 C -0.106
51 I 0.784
52 A -0.556
53 K -0.008
54 K -0.559
55 A -0.843
56 L -0.205
57 E -0.979
58 G -0.935
59 K -0.677
60 E -0.570
61 G -0.698
62 S -0.575
63 M 0.282
64 E -0.521
65 N -0.304
66 E 2.440
67 I 0.227
68 A -0.734
69 V 0.830
70 L 1.155
71 H -0.464
72 K -0.619
73 I 0.297
74 K -0.614
75 H -0.080
76 P -0.642
77 N 0.617
78 I 0.954
79 V 0.759
80 A -0.775
81 L 0.545
82 D -0.790
83 D 0.087
84 I -0.690
85 Y 0.121
86 E -0.093
87 S -0.157
88 G -0.952
89 G -1.094
90 H -0.950
91 L -0.121
92 Y -0.302
93 L 0.641
94 I 0.168
95 M 0.464
96 Q 0.632
97 L 0.095
98 V 0.272
99 S -0.760
100 G 2.059
101 G 1.435
102 E 1.615
103 L 1.716
104 F 1.908
105 D 0.968
106 R 0.134
107 I 1.438
108 V 0.147
109 E -0.550
110 K 0.132
111 G -0.046
112 F -0.636
113 Y 1.692
114 T 0.589
115 E 2.440
116 R -0.303
117 D 0.106
118 A 1.512
119 S 0.374
120 R -1.018
121 L 0.021
122 I -0.302
123 F -0.487
124 Q -0.829
125 V 0.607
126 L -0.036
127 D -0.774
128 A -0.009
129 V 0.350
130 K -0.931
131 Y -0.761
132 L 0.407
133 H 1.730
134 D -0.914
135 L -0.737
136 G -0.087
137 I 1.349
138 V 0.136
139 H 1.730
140 R 1.730
141 D 1.727
142 L 1.483
143 K 2.062
144 P 2.060
145 E 2.059
146 N 2.059
147 L 1.035
148 L 1.470
149 Y 0.374
150 Y -1.042
151 S -0.015
152 L -1.058
153 D -0.896
154 E -0.496
155 D -0.335
156 S 0.224
157 K -0.945
158 I 0.590
159 M 0.411
160 I 0.213
161 S 0.477
162 D 2.440
163 F 2.059
164 G 1.727
165 L 0.940
166 S 1.346
167 K -0.415
168 M -0.625
169 E -0.806
170 D -0.705
171 P -0.845
172 G -0.430
173 S -0.786
174 V -1.137
175 L -0.131
176 S -1.038
177 T -0.017
178 A -0.472
179 C 1.111
180 G 2.059
181 T 1.336
182 P 1.384
183 G 0.233
184 Y 2.440
185 V 0.399
186 A 1.199
187 P 2.060
188 E 1.621
189 V 1.242
190 L 0.988
191 A -0.784
192 Q -0.205
193 K -0.745
194 P -0.846
195 Y 1.520
196 S 0.155
197 K -0.733
198 A -0.022
199 V 0.274
200 D 2.060
201 C -0.037
202 W 2.062
203 S 1.042
204 I 0.163
205 G 2.440
206 V 1.443
207 I 1.308
208 A 0.122
209 Y 1.753
210 I 1.112
211 L 1.563
212 L 1.736
213 C 0.343
214 G 2.440
215 Y -0.065
216 P 1.118
217 P 2.440
218 F 2.440
219 Y 0.151
220 D 0.183
221 E 0.667
222 N -0.089
223 D -0.123
224 A -1.054
225 K -0.815
226 L 0.525
227 F 0.399
228 E -0.722
229 Q -0.432
230 I 1.376
231 L -0.289
232 K -0.840
233 A 0.135
234 E -0.946
235 Y 0.198
236 E -0.441
237 F 0.845
238 D -0.807
239 S -0.491
240 P -0.227
241 Y -0.015
242 W 2.008
243 D 0.001
244 D -0.936
245 I 1.272
246 S 1.194
247 D -0.771
248 S -0.803
249 A 1.736
250 K 1.510
251 D -0.162
252 F 0.530
253 I 1.008
254 R -0.801
255 H -0.499
256 L 0.829
257 M 1.066
258 E -0.834
259 K -0.883
260 D -0.307
261 P -0.186
262 E -0.839
263 K -0.830
264 R 2.440
265 F -0.532
266 T 0.074
267 C 0.337
268 E -0.938
269 Q 0.251
270 A 0.498
271 L 0.659
272 Q -0.658
273 H 1.024
274 P -0.263
275 W 1.587
276 I 0.272
277 A -0.927
278 G -0.751
279 D -0.803
280 T -0.941
281 A -0.633
282 L -1.054
283 D -0.818
284 K -0.968
285 N -0.977
286 I -0.600
287 H -0.808
288 Q -1.055
289 S -0.710
290 V 0.128
291 S -0.713
292 E -0.969
293 Q -0.594
294 I 0.519
295 K 0.128
296 K -0.532
297 N 0.106
298 F -0.478
299 A -0.665
300 K 0.020
301 S -0.403
302 K -0.188
303 W 0.132
304 K 0.891
305 Q -0.124
306 A -0.424
307 F -0.564
308 N -0.925
309 A -0.669
310 T 0.049
311 A -0.539
312 V -0.379
313 V -0.629
314 R -1.061
315 H -0.881
316 M -0.472
317 R -0.858
318 K -0.953
319 L -1.095
320 Q -0.989
321 L -1.123
322 G -0.941
323 T -1.170
324 S -1.000
325 Q -1.066
326 E -0.887
327 G -1.094
328 Q -1.046
329 G -1.097
330 Q -1.178
331 T -1.036
332 A -1.009
333 S -0.966
334 H -1.086
335 G -1.115
336 E -0.952
337 L -1.132
338 L -1.066
339 T -0.999
340 P -1.023
341 T -1.056
342 A -0.956
343 G -1.036
344 G -1.023
345 P -0.945
346 A -1.097
347 A -0.924
348 G -1.179
349 C -0.952
350 C -1.133
351 C -1.127
352 R -0.986
353 D -0.982
354 C -1.078
355 C -1.054
356 V -1.215
357 E -0.964
358 P -1.076
359 G -0.912
360 S -0.965
361 E -0.992
362 L -1.052
363 P -1.000 *
364 P -1.000 *
365 A -1.000 *
366 P -1.000 *
367 P -1.000 *
368 P -1.000 *
369 S -1.000 *
370 S -1.000 *
371 R -1.000 *
372 A -1.000 *
373 M -1.000 *
374 D -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 143.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MPGAVEGPRW KQAEDIRDIY DFRDVLGTGA FSEVILAEDK RTQKLVAIKC IAKKALEGKE
70 80 90 100 110 120
GSMENEIAVL HKIKHPNIVA LDDIYESGGH LYLIMQLVSG GELFDRIVEK GFYTERDASR
130 140 150 160 170 180
LIFQVLDAVK YLHDLGIVHR DLKPENLLYY SLDEDSKIMI SDFGLSKMED PGSVLSTACG
190 200 210 220 230 240
TPGYVAPEVL AQKPYSKAVD CWSIGVIAYI LLCGYPPFYD ENDAKLFEQI LKAEYEFDSP
250 260 270 280 290 300
YWDDISDSAK DFIRHLMEKD PEKRFTCEQA LQHPWIAGDT ALDKNIHQSV SEQIKKNFAK
310 320 330 340 350 360
SKWKQAFNAT AVVRHMRKLQ LGTSQEGQGQ TASHGELLTP TAGGPAAGCC CRDCCVEPGS
370
ELPPAPPPSS RAMD
1A06 (X-Ray,2.50 Å resolution)
1MXE (X-Ray,1.70 Å resolution)
CaMKI has been implicated in several cellular processes. CaMKI has multiple isoforms. Stedman et al. showed that CaMKIα has a functional NES overlapping the CaM-binding domain. Preincubation of CaMKIα with Ca2+/CaM increased binding of CaMKIα to CRM1. It was possibly due to the conformational change due to Ca2+/CaM binding.
[1]. "Cytoplasmic localization of calcium/calmodulin-dependent protein kinase I-alpha depends on a nuclear export signal in its regulatory domain"
Stedman DR, Uboha NV, Stedman TT, Nairn AC, Picciotto MR. (2004)
FEBS Lett,
566:275-280
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
