Summary for FLIP-L (NES ID: 147)
Full Name
CASP8 and FADD-like apoptosis regulator
UniProt Alternative Names
Cellular FLICE-like inhibitory protein (c-FLIP)
Caspase-eight-related protein (Casper)
Caspase-like apoptosis regulatory protein (CLARP)
MACH-related inducer of toxicity (MRIT)
Caspase homolog (CASH)
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
Undetermined
Secondary Structure of Export Signal
α-helix (residues 439-446)
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|12643547|sp|O15519.1|CFLAR_HUMAN RecName: Full=CASP8 and FADD-like apoptosis regulator; AltName: Full=Caspase homolog; Short=CASH; AltName: Full=Caspase-eight-related protein; Short=Casper; AltName: Full=Caspase-like apoptosis regulatory protein; Short=CLARP; AltName: Full=Cellular FLICE-like inhibitory protein; Short=c-FLIP; AltName: Full=FADD-like antiapoptotic molecule 1; Short=FLAME-1; AltName: Full=Inhibitor of FLICE; Short=I-FLICE; AltName: Full=MACH-related inducer of toxicity; Short=MRIT; AltName: Full=Usurpin; Contains: RecName: Full=CASP8 and FADD-like apoptosis regulator subunit p43; Contains: RecName: Full=CASP8 and FADD-like apoptosis regulator subunit p12; Flags: Precursor
MSAEVIHQVEEALDTDEKEMLLFLCRDVAIDVVPPNVRDLLDILRERGKLSVGDLAELLYRVRRFDLLKR
ILKMDRKAVETHLLRNPHLVSDYRVLMAEIGEDLDKSDVSSLIFLMKDYMGRGKISKEKSFLDLVVELEK
LNLVAPDQLDLLEKCLKNIHRIDLKTKIQKYKQSVQGAGTSYRNVLQAAIQKSLKDPSNNFRLHNGRSKE
QRLKEQLGAQQEPVKKSIQESEAFLPQSIPEERYKMKSKPLGICLIIDCIGNETELLRDTFTSLGYEVQK
FLHLSMHGISQILGQFACMPEHRDYDSFVCVLVSRGGSQSVYGVDQTHSGLPLHHIRRMFMGDSCPYLAG
KPKMFFIQNYVVSEGQLEDSSLLEVDGPAMKNVEFKAQKRGLCTVHREADFFWSLCTADMSLLEQSHSSP
SLYLQCLSQKLRQERKRPLLDLHIELNGYMYDWNSRVSAKEKYYVWLQHTLRKKLILSYT
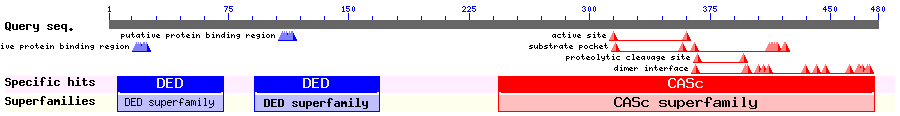
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 984899999960899689986742138688989989899999999929999489888986
Pred: CCHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHCCCCCCCCHHHHHH
AA: MSAEVIHQVEEALDTDEKEMLLFLCRDVAIDVVPPNVRDLLDILRERGKLSVGDLAELLY
10 20 30 40 50 60
Conf: 418078999985269899986204899865734787898641899211102021013745
Pred: HHCHHHHHHHHHCCCHHHHHHHCCCCCCCCCHHHHHHHHHHCCCCHHHHHHHHHCCCCCC
AA: RVRRFDLLKRILKMDRKAVETHLLRNPHLVSDYRVLMAEIGEDLDKSDVSSLIFLMKDYM
70 80 90 100 110 120
Conf: 675567988865899985450698999925999987015910279999997611048998
Pred: CCCCCCCCCCHHHHHHHHHHHCCCCCCCHHHHHHHHHHCCCHHHHHHHHHHHHHHCCCCC
AA: GRGKISKEKSFLDLVVELEKLNLVAPDQLDLLEKCLKNIHRIDLKTKIQKYKQSVQGAGT
130 140 150 160 170 180
Conf: 653124566411579999843347887300244532024679876542222246899987
Pred: CCCCHHHHHHHCCCCCCCCCCCCCCCCCCHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCC
AA: SYRNVLQAAIQKSLKDPSNNFRLHNGRSKEQRLKEQLGAQQEPVKKSIQESEAFLPQSIP
190 200 210 220 230 240
Conf: 532004888604899955964114556310224772899977999899999999880898
Pred: CCCCCCCCCCCEEEEEECCCCCCHHHHHHHCCCCCCEEEEECCCCHHHHHHHHHHHHCCC
AA: EERYKMKSKPLGICLIIDCIGNETELLRDTFTSLGYEVQKFLHLSMHGISQILGQFACMP
250 260 270 280 290 300
Conf: 999998258999701599928875599998444666105679999863499951177430
Pred: CCCCCCEEEEEEEECCCCCCEEEECCCCCCCCHHHHHCCCCCCCCCCCCCCCCEEEEECC
AA: EHRDYDSFVCVLVSRGGSQSVYGVDQTHSGLPLHHIRRMFMGDSCPYLAGKPKMFFIQNY
310 320 330 340 350 360
Conf: 441788788862025787776223542028998899431110020302883641339999
Pred: EECCCCCCCCCCCCCCCCCCCCHHHHHHCCCCCCCCCCCCEEEECCCCCCEEEEECCCCC
AA: VVSEGQLEDSSLLEVDGPAMKNVEFKAQKRGLCTVHREADFFWSLCTADMSLLEQSHSSP
370 380 390 400 410 420
Conf: 203999999985005997368999862000012478899984110003200241013589
Pred: CHHHHHHHHHHHHCCCCCHHHHHHHHHHHHCCCCCCCCCCCCEEECCCCCCCCCCCCCCC
AA: SLYLQCLSQKLRQERKRPLLDLHIELNGYMYDWNSRVSAKEKYYVWLQHTLRKKLILSYT
430 440 450 460 470 480
Conf:
Pred:
AA:
Show Conservation Score by AL2CO
1 M -1.000 *
2 S -1.000 *
3 A -1.000 *
4 E -1.000 *
5 V -1.000 *
6 I -1.000 *
7 H -1.000 *
8 Q -1.000 *
9 V -1.000 *
10 E -1.000 *
11 E -1.000 *
12 A -1.000 *
13 L -1.000 *
14 D -1.000 *
15 T -1.000 *
16 D -1.000 *
17 E -1.000 *
18 K -1.000 *
19 E -1.000 *
20 M -1.000 *
21 L -1.000 *
22 L -1.000 *
23 F -1.000 *
24 L -1.000 *
25 C -1.000 *
26 R -1.000 *
27 D -1.000 *
28 V -1.000 *
29 A -1.000 *
30 I -1.000 *
31 D -1.000 *
32 V -1.000 *
33 V -1.000 *
34 P -1.000 *
35 P -1.000 *
36 N -1.000 *
37 V -1.000 *
38 R -1.000 *
39 D -1.000 *
40 L -1.000 *
41 L -1.000 *
42 D -1.000 *
43 I -1.000 *
44 L -1.000 *
45 R -1.000 *
46 E -1.000 *
47 R -1.000 *
48 G -1.000 *
49 K -1.000 *
50 L -1.000 *
51 S -1.000 *
52 V -1.000 *
53 G -1.000 *
54 D -1.000 *
55 L -1.000 *
56 A -1.000 *
57 E -1.000 *
58 L -1.000 *
59 L -1.000 *
60 Y -1.000 *
61 R -1.000 *
62 V -1.000 *
63 R -1.000 *
64 R -1.000 *
65 F -1.000 *
66 D -1.000 *
67 L -1.000 *
68 L -1.000 *
69 K -1.000 *
70 R -1.000 *
71 I -1.000 *
72 L -1.000 *
73 K -1.000 *
74 M -1.000 *
75 D -1.000 *
76 R -1.000 *
77 K -1.000 *
78 A -1.000 *
79 V -1.000 *
80 E -1.000 *
81 T -1.000 *
82 H -1.000 *
83 L -1.000 *
84 L -1.000 *
85 R -1.000 *
86 N -1.000 *
87 P -1.000 *
88 H -1.000 *
89 L -1.000 *
90 V -1.000 *
91 S -1.000 *
92 D -0.272
93 Y 1.392
94 R 1.859
95 V -0.148
96 L 0.932
97 M 1.609
98 A 0.221
99 E -0.005
100 I 1.792
101 G 0.326
102 E 1.091
103 D 0.114
104 L 1.071
105 D -0.128
106 K -0.204
107 S -0.246
108 D 0.088
109 V 0.883
110 S -0.179
111 S -0.427
112 L 0.442
113 I 0.237
114 F 0.987
115 L 0.712
116 M -0.103
117 K -0.804
118 D -0.239
119 Y -0.682
120 M 0.661
121 G 0.248
122 R 0.378
123 G -0.501
124 K -0.233
125 I -0.277
126 S -0.521
127 K -0.378
128 E -0.662
129 K -0.573
130 S 0.625
131 F -0.310
132 L 0.530
133 D -0.047
134 L 0.683
135 V 0.698
136 V -0.407
137 E -0.300
138 L 1.720
139 E 1.650
140 K 0.386
141 L -0.414
142 N -0.448
143 L 0.005
144 V 0.493
145 A -0.173
146 P 0.034
147 D -0.304
148 Q -0.100
149 L 0.515
150 D -0.602
151 L -0.632
152 L 0.970
153 E -0.349
154 K -0.410
155 C 0.215
156 L 0.851
157 K -0.690
158 N -0.549
159 I -0.118
160 H -0.626
161 R -0.191
162 I -0.924
163 D -0.505
164 L 0.593
165 K -0.346
166 T -0.511
167 K -0.437
168 I 1.362
169 Q -0.500
170 K -0.644
171 Y -0.441
172 K -0.343
173 Q -0.692
174 S -0.665
175 V -0.821
176 Q -0.747
177 G -0.656
178 A -0.875
179 G -0.936
180 T -0.719
181 S -0.801
182 Y -1.049
183 R -0.737
184 N -0.908
185 V -0.822
186 L -0.928
187 Q -0.726
188 A -0.713
189 A -0.957
190 I -0.949
191 Q -0.897
192 K -0.886
193 S -0.668
194 L -0.886
195 K -0.920
196 D -0.879
197 P -0.973
198 S -0.908
199 N -0.951
200 N -0.793
201 F -1.019
202 R -0.889
203 L -1.029
204 H -0.994
205 N -1.055
206 G -0.797
207 R -0.993
208 S -0.870
209 K -0.877
210 E -0.849
211 Q -0.742
212 R -0.988
213 L -1.031
214 K -0.900
215 E -0.726
216 Q -0.956
217 L -1.009
218 G -0.878
219 A -0.944
220 Q -0.944
221 Q -0.871
222 E -0.921
223 P -0.951
224 V -0.927
225 K -0.893
226 K -0.899
227 S -0.729
228 I -0.878
229 Q -0.833
230 E -0.827
231 S -0.856
232 E -0.764
233 A -0.799
234 F -0.936
235 L -1.044
236 P -0.886
237 Q -0.891
238 S -1.003
239 I -0.964
240 P -1.012
241 E -0.825
242 E -0.826
243 R -0.901
244 Y 2.543
245 K -0.344
246 M 0.607
247 K -0.194
248 S -0.490
249 K -0.589
250 P -0.221
251 L 0.279
252 G 1.984
253 I -0.596
254 C 0.988
255 L 0.590
256 I 1.615
257 I 1.052
258 D 0.935
259 C 0.233
260 I -0.572
261 G -0.639
262 N -0.387
263 E 0.238
264 T -0.760
265 E -0.645
266 L -0.879
267 L -0.358
268 R -0.780
269 D -0.451
270 T 0.547
271 F 2.075
272 T -0.478
273 S -0.709
274 L 1.611
275 G 0.066
276 Y 2.002
277 E -0.404
278 V 1.105
279 Q -0.695
280 K -0.882
281 F -0.456
282 L -0.711
283 H 0.226
284 L -0.172
285 S -0.247
286 M -0.685
287 H -0.646
288 G -0.334
289 I 1.001
290 S -0.879
291 Q -0.650
292 I -0.563
293 L 0.852
294 G -0.865
295 Q -0.672
296 F -0.093
297 A 0.196
298 C -0.502
299 M -0.652
300 P -0.668
301 E -0.192
302 H 0.249
303 R -0.580
304 D -0.515
305 Y -0.807
306 D 1.317
307 S 1.198
308 F 1.387
309 V 0.755
310 C 1.099
311 V 0.834
312 L 0.897
313 V 1.291
314 S 2.446
315 R 2.187
316 G 3.332
317 G -0.633
318 S -0.732
319 Q 0.341
320 S -0.793
321 V 1.301
322 Y 0.153
323 G 1.287
324 V -0.257
325 D 1.973
326 Q -0.475
327 T -0.687
328 H -0.667
329 S -1.000 *
330 G -0.461
331 L 0.889
332 P -0.490
333 L 0.811
334 H -0.153
335 H -0.768
336 I 1.244
337 R -0.087
338 R -0.572
339 M -0.690
340 F 1.748
341 M -0.243
342 G 0.918
343 D -0.691
344 S -0.702
345 C 1.388
346 P -0.273
347 Y -0.370
348 L 2.970
349 A -0.279
350 G -0.035
351 K 2.985
352 P 3.805
353 K 2.993
354 M 1.415
355 F 2.748
356 F 1.810
357 I 0.838
358 Q 2.234
359 N 0.675
360 Y 2.414
361 V 0.034
362 V 0.571
363 S -0.585
364 E -0.271
365 G -0.775
366 Q -0.716
367 L -0.971
368 E -0.625
369 D -0.818
370 S -0.663
371 S -0.608
372 L -1.001
373 L -0.610
374 E -0.377
375 V -0.695
376 D -0.130
377 G -0.479
378 P -0.753
379 A -0.797
380 M -0.937
381 K -0.783
382 N -0.912
383 V -0.925
384 E -0.865
385 F -0.819
386 K -0.894
387 A -0.634
388 Q -0.877
389 K -0.921
390 R -0.921
391 G -0.932
392 L -0.995
393 C -0.976
394 T -0.570
395 V 1.362
396 H 2.089
397 R -0.692
398 E -0.703
399 A 1.540
400 D 3.805
401 F 0.868
402 F 1.811
403 W 0.231
404 S 1.086
405 L 0.272
406 C 1.499
407 T 0.242
408 A 0.349
409 D -0.053
410 M -0.206
411 S 0.089
412 L -0.378
413 L 1.318
414 E -0.161
415 Q 1.355
416 S -0.069
417 H -0.426
418 S -0.878
419 S -0.634
420 P 0.629
421 S 2.186
422 L 0.083
423 Y 2.442
424 L 1.350
425 Q 0.176
426 C -0.377
427 L 1.630
428 S 0.328
429 Q -0.410
430 K -0.626
431 L 1.600
432 R -0.642
433 Q -0.566
434 E -0.640
435 R -0.159
436 K -0.250
437 R -0.668
438 P 0.354
439 L 1.255
440 L 0.208
441 D -0.022
442 L 1.129
443 H 1.782
444 I 0.840
445 E -0.756
446 L 2.450
447 N 0.008
448 G -0.383
449 Y -0.561
450 M 1.260
451 Y -0.061
452 D -0.099
453 W -0.589
454 N -0.800
455 S -0.501
456 R -0.843
457 V -0.839
458 S -0.827
459 A -0.744
460 K -0.530
461 E -0.605
462 K 0.687
463 Y 1.591
464 Y -0.183
465 V 0.615
466 W -0.523
467 L 0.153
468 Q -0.590
469 H 0.349
470 T 0.727
471 L 3.081
472 R 1.355
473 K 1.772
474 K -0.115
475 L 1.405
476 I 0.090
477 L 2.673
478 S 0.484
479 Y -1.000 *
480 T -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 147.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MSAEVIHQVE EALDTDEKEM LLFLCRDVAI DVVPPNVRDL LDILRERGKL SVGDLAELLY
70 80 90 100 110 120
RVRRFDLLKR ILKMDRKAVE THLLRNPHLV SDYRVLMAEI GEDLDKSDVS SLIFLMKDYM
130 140 150 160 170 180
GRGKISKEKS FLDLVVELEK LNLVAPDQLD LLEKCLKNIH RIDLKTKIQK YKQSVQGAGT
190 200 210 220 230 240
SYRNVLQAAI QKSLKDPSNN FRLHNGRSKE QRLKEQLGAQ QEPVKKSIQE SEAFLPQSIP
250 260 270 280 290 300
EERYKMKSKP LGICLIIDCI GNETELLRDT FTSLGYEVQK FLHLSMHGIS QILGQFACMP
310 320 330 340 350 360
EHRDYDSFVC VLVSRGGSQS VYGVDQTHSG LPLHHIRRMF MGDSCPYLAG KPKMFFIQNY
370 380 390 400 410 420
VVSEGQLEDS SLLEVDGPAM KNVEFKAQKR GLCTVHREAD FFWSLCTADM SLLEQSHSSP
430 440 450 460 470 480
SLYLQCLSQK LRQERKRPLL DLHIELNGYM YDWNSRVSAK EKYYVWLQHT LRKKLILSYT
3D Structures in PDB
3H11 (X-Ray,1.90 Å resolution)
Comments
cFLIP-L is a cytoplasmic protein that inhibits the apoptosis signaling initiated by death receptor ligation at the cell membrane. cFLIP-L can also enhance Wnt signaling by inhibiting ubiquitylation of &beta-catenin, a mediator of Wnt signaling. Katayama et al. showed that cFLIP-L is present in both the nucleus and the cytoplasm and there are both a bipartie NLS and NES present at its C-terminus. NLS- and NES-mutants can only marginally enhance Wnt signaling as compared to the wild type, although the anti-apoptotic function was not apparently affected by these mutations.
Doubts about NESs: The proposed NES failed to bind CRM1 in GST-pulldown assay (Chook Lab, unpublished results).
References
[1]. "Modulation of Wnt signaling by the nuclear localization of cellular FLIP-L."
Katayama R, Ishioka T, Takada S, Takada R, Fujita N, Tsuruo T, Naito M (2010)
J Cell Sci,
123:23-8
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
