Mitogen-activated protein kinase phosphatase 3
UniProt
Dual specificity protein phosphatase 6, Dual specificity protein phosphatase PYST1, MAPK-3
Show FASTA Format
>gi|108860971|sp|Q16828.2|DUS6_HUMAN RecName: Full=Dual specificity protein phosphatase 6; AltName: Full=Dual specificity protein phosphatase PYST1; AltName: Full=Mitogen-activated protein kinase phosphatase 3; Short=MAP kinase phosphatase 3; Short=MKP-3
MIDTLRPVPFASEMAISKTVAWLNEQLELGNERLLLMDCRPQELYESSHIESAINVAIPGIMLRRLQKGN
LPVRALFTRGEDRDRFTRRCGTDTVVLYDESSSDWNENTGGESVLGLLLKKLKDEGCRAFYLEGGFSKFQ
AEFSLHCETNLDGSCSSSSPPLPVLGLGGLRISSDSSSDIESDLDRDPNSATDSDGSPLSNSQPSFPVEI
LPFLYLGCAKDSTNLDVLEEFGIKYILNVTPNLPNLFENAGEFKYKQIPISDHWSQNLSQFFPEAISFID
EARGKNCGVLVHCLAGISRSVTVTVAYLMQKLNLSMNDAYDIVKMKKSNISPNFNFMGQLLDFERTLGLS
SPCDNRVPAQQLYFTTPSNQNVYQVDSLQST
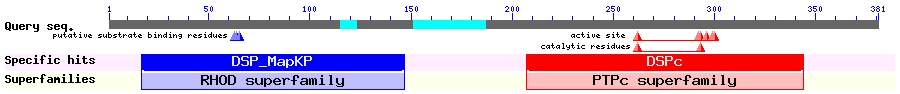
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 988888852012444432099999998228997599814782111001344543447636
Pred: CCCCCCCCCCHHHHHHHHHHHHHHHHHHHCCCCEEEEECCCCHHHHCCCCCCCCCCCCCH
AA: MIDTLRPVPFASEMAISKTVAWLNEQLELGNERLLLMDCRPQELYESSHIESAINVAIPG
10 20 30 40 50 60
Conf: 789763189867421124752011010148986089727998656568899535999999
Pred: HHHHHHCCCCCCCCCCCCCCCCHHHHCCCCCCCEEEEECCCCCCCCCCCCCCCHHHHHHH
AA: IMLRRLQKGNLPVRALFTRGEDRDRFTRRCGTDTVVLYDESSSDWNENTGGESVLGLLLK
70 80 90 100 110 120
Conf: 966609612231430554885302122247799989999998744578853358999875
Pred: HHHHCCCEEEECCCCHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: KLKDEGCRAFYLEGGFSKFQAEFSLHCETNLDGSCSSSSPPLPVLGLGGLRISSDSSSDI
130 140 150 160 170 180
Conf: 677789999998999999999999999233343102585466887689852991999719
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCEEECCCCCCCCCCCCCCHHHHHHCCCEEEEECC
AA: ESDLDRDPNSATDSDGSPLSNSQPSFPVEILPFLYLGCAKDSTNLDVLEEFGIKYILNVT
190 200 210 220 230 240
Conf: 999986546898706796058864665000026888879999847990889508986503
Pred: CCCCCCCCCCCCCEEEEEECCCCCCCCCCCCHHHHHHHHHHHHHCCCEEEEEECCCCCCH
AA: PNLPNLFENAGEFKYKQIPISDHWSQNLSQFFPEAISFIDEARGKNCGVLVHCLAGISRS
250 260 270 280 290 300
Conf: 589999998623889688999986118987899413555699999847999999999998
Pred: HHHHHHHHHHHCCCCHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHCCCCCCCCCCCCC
AA: VTVTVAYLMQKLNLSMNDAYDIVKMKKSNISPNFNFMGQLLDFERTLGLSSPCDNRVPAQ
310 320 330 340 350 360
Conf: 630026888760115998889
Pred: CCCCCCCCCCCCEECCCCCCC
AA: QLYFTTPSNQNVYQVDSLQST
370 380
Show Conservation Score by AL2CO
1 M -1.000 *
2 I -1.000 *
3 D -1.000 *
4 T -1.000 *
5 L -1.000 *
6 R -1.000 *
7 P -1.000 *
8 V -1.000 *
9 P -1.000 *
10 F -1.000 *
11 A -1.000 *
12 S -1.000 *
13 E -1.000 *
14 M -1.000 *
15 A -1.000 *
16 I -1.000 *
17 S -1.000 *
18 K -1.000 *
19 T -1.000 *
20 V -1.000 *
21 A -1.000 *
22 W -1.000 *
23 L 1.457
24 N -0.392
25 E -0.203
26 Q -0.482
27 L 0.109
28 E -0.308
29 L -0.735
30 G -0.448
31 N -0.474
32 E -0.510
33 R -0.441
34 L -0.005
35 L 0.570
36 L 1.605
37 M 1.762
38 D 3.762
39 C 0.622
40 R 3.124
41 P 0.113
42 Q -0.466
43 E -0.810
44 L -0.238
45 Y 1.398
46 E -0.223
47 S -0.478
48 S -0.259
49 H 0.920
50 I 1.832
51 E -0.768
52 S -0.232
53 A 2.206
54 I -0.348
55 N 0.803
56 V 0.670
57 A -0.570
58 I -0.198
59 P 0.214
60 G -0.298
61 I 0.060
62 M -0.180
63 L -0.124
64 R 0.784
65 R 0.211
66 L -0.146
67 Q -0.509
68 K -0.503
69 G -0.167
70 N -0.441
71 L -0.012
72 P -0.445
73 V -0.451
74 R -0.716
75 A -0.777
76 L 0.071
77 F -0.040
78 T -0.561
79 R -0.601
80 G -0.594
81 E -0.634
82 D -0.669
83 R -0.283
84 D -0.711
85 R -0.556
86 F -0.537
87 T -0.740
88 R -0.565
89 R -0.692
90 C -0.894
91 G -0.762
92 T -0.878
93 D -0.620
94 T -0.801
95 V 0.379
96 V 0.760
97 L 0.611
98 Y 0.306
99 D 0.694
100 E -0.024
101 S -0.519
102 S -0.246
103 S -0.620
104 D -0.439
105 W -0.858
106 N -0.577
107 E -0.792
108 N -0.752
109 T -0.685
110 G -0.586
111 G -0.622
112 E -0.686
113 S -0.629
114 V -0.787
115 L -0.356
116 G -0.900
117 L -0.693
118 L -0.115
119 L -0.541
120 K -0.582
121 K -0.661
122 L -0.161
123 K -0.846
124 D -0.589
125 E -0.848
126 G -0.659
127 C -0.873
128 R -0.498
129 A -0.265
130 F -0.767
131 Y -0.477
132 L 0.060
133 E -0.601
134 G 0.030
135 G 1.091
136 F 0.208
137 S -0.614
138 K -0.356
139 F 0.290
140 Q -0.629
141 A -0.620
142 E -0.827
143 F -0.135
144 S -0.166
145 L -0.526
146 H -0.655
147 C -0.125
148 E -0.818
149 T -0.478
150 N -0.776
151 L -0.868
152 D -0.693
153 G -0.896
154 S -0.881
155 C -0.577
156 S -0.890
157 S -0.930
158 S -0.745
159 S -0.909
160 P -0.834
161 P -0.539
162 L -0.730
163 P -0.642
164 V -0.717
165 L -0.781
166 G -0.833
167 L -0.880
168 G -0.797
169 G -0.699
170 L -0.759
171 R -0.841
172 I -0.816
173 S -0.938
174 S -0.676
175 D -0.861
176 S -0.865
177 S -0.823
178 S -0.827
179 D -0.803
180 I -0.876
181 E -0.621
182 S -0.787
183 D -0.785
184 L -0.859
185 D -0.732
186 R -0.665
187 D -0.719
188 P -0.596
189 N -0.698
190 S -0.737
191 A -0.832
192 T -0.866
193 D -0.696
194 S -0.887
195 D -0.922
196 G -0.908
197 S -0.729
198 P -0.384
199 L -0.873
200 S -0.686
201 N -0.661
202 S -0.775
203 Q -0.842
204 P -1.000 *
205 S -0.637
206 F -0.628
207 P 1.147
208 V 0.054
209 E -0.102
210 I 2.436
211 L 0.349
212 P 0.453
213 F -0.176
214 L 2.119
215 Y 1.645
216 L 1.520
217 G 2.563
218 C 0.372
219 A -0.521
220 K -0.726
221 D 0.441
222 S 1.648
223 T -0.537
224 N 0.109
225 L -0.707
226 D -0.453
227 V -0.627
228 L 0.375
229 E -0.471
230 E -0.513
231 F -0.155
232 G 0.163
233 I 1.300
234 K 0.108
235 Y 0.283
236 I 0.866
237 L 1.932
238 N 1.383
239 V 0.876
240 T 0.478
241 P -0.680
242 N -0.329
243 L -0.521
244 P -0.522
245 N -0.595
246 L -0.770
247 F -0.431
248 E -0.564
249 N -0.892
250 A -0.905
251 G -0.492
252 E -0.646
253 F -0.257
254 K -0.508
255 Y 0.582
256 K 0.395
257 Q -0.417
258 I 1.562
259 P -0.234
260 I 1.278
261 S -0.534
262 D 3.489
263 H -0.239
264 W -0.812
265 S -0.594
266 Q 0.143
267 N 0.546
268 L 1.993
269 S -0.326
270 Q -0.558
271 F -0.028
272 F 1.412
273 P -0.556
274 E -0.382
275 A 0.637
276 I -0.196
277 S -0.209
278 F 1.061
279 I 1.448
280 D -0.053
281 E -0.588
282 A -0.088
283 R -0.430
284 G -0.598
285 K -0.535
286 N 0.027
287 C -0.240
288 G 0.009
289 V 1.343
290 L 1.610
291 V 2.281
292 H 1.729
293 C 2.132
294 L -0.616
295 A 0.102
296 G 3.694
297 I 0.928
298 S 3.703
299 R 1.959
300 S 2.184
301 V 0.749
302 T 1.554
303 V 1.166
304 T 1.009
305 V 1.132
306 A 0.982
307 Y 2.144
308 L 1.520
309 M 2.207
310 Q -0.476
311 K -0.430
312 L -0.506
313 N -0.318
314 L 0.271
315 S 0.006
316 M 0.527
317 N -0.616
318 D 0.012
319 A 2.161
320 Y 0.339
321 D -0.134
322 I -0.124
323 V 1.030
324 K 0.378
325 M -0.309
326 K 0.077
327 K 1.660
328 S -0.443
329 N -0.348
330 I 0.988
331 S -0.342
332 P 3.635
333 N 2.425
334 F -0.677
335 N 0.831
336 F 2.750
337 M 0.431
338 G -0.678
339 Q 1.403
340 L 4.126
341 L -0.444
342 D -0.154
343 F 0.474
344 E 1.726
345 R -0.391
346 T -0.633
347 L 1.534
348 G -0.710
349 L -0.531
350 S -0.503
351 S -0.642
352 P -0.574
353 C -0.588
354 D -1.000 *
355 N -1.000 *
356 R -1.000 *
357 V -1.000 *
358 P -1.000 *
359 A -1.000 *
360 Q -1.000 *
361 Q -1.000 *
362 L -1.000 *
363 Y -1.000 *
364 F -1.000 *
365 T -1.000 *
366 T -1.000 *
367 P -1.000 *
368 S -1.000 *
369 N -1.000 *
370 Q -1.000 *
371 N -1.000 *
372 V -1.000 *
373 Y -1.000 *
374 Q -1.000 *
375 V -1.000 *
376 D -1.000 *
377 S -1.000 *
378 L -1.000 *
379 Q -1.000 *
380 S -1.000 *
381 T -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 163.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MIDTLRPVPF ASEMAISKTV AWLNEQLELG NERLLLMDCR PQELYESSHI ESAINVAIPG
70 80 90 100 110 120
IMLRRLQKGN LPVRALFTRG EDRDRFTRRC GTDTVVLYDE SSSDWNENTG GESVLGLLLK
130 140 150 160 170 180
KLKDEGCRAF YLEGGFSKFQ AEFSLHCETN LDGSCSSSSP PLPVLGLGGL RISSDSSSDI
190 200 210 220 230 240
ESDLDRDPNS ATDSDGSPLS NSQPSFPVEI LPFLYLGCAK DSTNLDVLEE FGIKYILNVT
250 260 270 280 290 300
PNLPNLFENA GEFKYKQIPI SDHWSQNLSQ FFPEAISFID EARGKNCGVL VHCLAGISRS
310 320 330 340 350 360
VTVTVAYLMQ KLNLSMNDAY DIVKMKKSNI SPNFNFMGQL LDFERTLGLS SPCDNRVPAQ
370 380
QLYFTTPSNQ NVYQVDSLQS T
MKP3 is one of the 10 distinct MKPs in mammalian cells. It preferentially dephosphorylates the stress-activated MAP kinases c-Jun amino-terminal kinase and p38. The NES is also conserved in MKP-4, whose export is LMB sensitive too.
[1]. "Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase MKP-3 and its ability to anchor MAP kinase in the cytoplasm are mediated by a conserved nuclear export signal."
Karlsson M, Mathers J, Dickinson RJ, Mandl M, Keyse SM. (2004)
J Biol Chem,
279:41882-91
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
