MAP kinase-activated protein kinase 5
UniProt
Show FASTA Format
>gi|52000764|sp|O54992.1|MAPK5_MOUSE RecName: Full=MAP kinase-activated protein kinase 5; Short=MAPK-activated protein kinase 5; Short=MAPKAP kinase 5; Short=MAPKAPK-5
MSEDSDMEKAIKETSILEEYSINWTQKLGAGISGPVRVCVKKSTQERFALKILLDRPKARNEVRLHMMCA
THPNIVQIIEVFANSVQFPHESSPRARLLIVMEMMEGGELFHRISQHRHFTEKQASQVTKQIALALQHCH
LLNIAHRDLKPENLLFKDNSLDAPVKLCDFGFAKVDQGDLMTPQFTPYYVAPQVLEAQRRHQKEKSGIIP
TSPTPYTYNKSCDLWSLGVIIYVMLCGYPPFYSKHHSRTIPKDMRKKIMTGSFEFPEEEWSQISEMAKDV
VRKLLKVKPEERLTIEGVLDHPWLNSTEALDNVLPSAQLMMDKAVVAGIQQAHAEQLANMRIQDLKVSLK
PLHSVNNPILRKRKLLGTKPKDGIYIHDHENGTEDSNVALEKLRDVIAQCILPQAGKGENEDEKLNEVMQ
EAWKYNRECKLLRDALQSFSWNGRGFTDKVDRLKLAEVVKQVIEEQTLPHEPQ
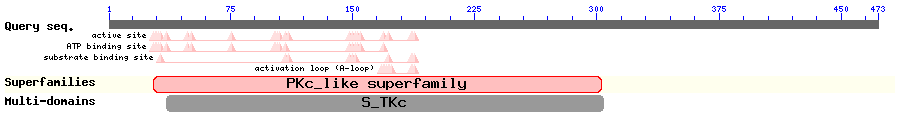
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 997425688632078522276068310023466351895644345613789833862328
Pred: CCCCHHHHHHHHCCCCCEEEECCCCCEECCCCCCCEEEEEECCCCCEEEEEEECCCCCCH
AA: MSEDSDMEKAIKETSILEEYSINWTQKLGAGISGPVRVCVKKSTQERFALKILLDRPKAR
10 20 30 40 50 60
Conf: 899999985199981269876225778999899983289998503765112433128999
Pred: HHHHHHHHHCCCCCCEEEEEEECCCCCCCCCCCCCCEEEEEEEECCCCCHHHHHHCCCCC
AA: NEVRLHMMCATHPNIVQIIEVFANSVQFPHESSPRARLLIVMEMMEGGELFHRISQHRHF
70 80 90 100 110 120
Conf: 978899999999999999875486345799975234679999980010134332258999
Pred: CHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCEEECCCCCCCCCCCC
AA: TEKQASQVTKQIALALQHCHLLNIAHRDLKPENLLFKDNSLDAPVKLCDFGFAKVDQGDL
130 140 150 160 170 180
Conf: 887557765627989998740022367677999998788866304266799988439999
Pred: CCCCCCCCCCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHCCCCC
AA: MTPQFTPYYVAPQVLEAQRRHQKEKSGIIPTSPTPYTYNKSCDLWSLGVIIYVMLCGYPP
190 200 210 220 230 240
Conf: 855899999966899966525445899703333299999997624889997766555422
Pred: CCCCCCCCCCCHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHC
AA: FYSKHHSRTIPKDMRKKIMTGSFEFPEEEWSQISEMAKDVVRKLLKVKPEERLTIEGVLD
250 260 270 280 290 300
Conf: 997678888998888356430046899999999999984130011123123556671123
Pred: CCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCHHH
AA: HPWLNSTEALDNVLPSAQLMMDKAVVAGIQQAHAEQLANMRIQDLKVSLKPLHSVNNPIL
310 320 330 340 350 360
Conf: 553201689999953235889887713456878999866148999986455489999999
Pred: HHHHHCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHH
AA: RKRKLLGTKPKDGIYIHDHENGTEDSNVALEKLRDVIAQCILPQAGKGENEDEKLNEVMQ
370 380 390 400 410 420
Conf: 98898665299999531214458776652479999999999987332999999
Pred: HHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHCCCCCCC
AA: EAWKYNRECKLLRDALQSFSWNGRGFTDKVDRLKLAEVVKQVIEEQTLPHEPQ
430 440 450 460 470
Show Conservation Score by AL2CO
1 M -1.000 *
2 S -1.000 *
3 E -1.000 *
4 D -1.000 *
5 S -1.000 *
6 D -1.000 *
7 M -1.000 *
8 E -1.000 *
9 K -1.000 *
10 A -1.000 *
11 I -1.000 *
12 K -1.000 *
13 E -0.746
14 T -0.935
15 S -0.910
16 I 0.232
17 L -0.904
18 E -0.666
19 E -0.965
20 Y 1.394
21 S -0.938
22 I -0.163
23 N -0.818
24 W -1.000 *
25 T -1.000 *
26 Q -0.155
27 K -0.393
28 L 1.250
29 G 2.249
30 A -0.726
31 G 2.618
32 I -0.767
33 S 0.247
34 G 1.510
35 P -0.456
36 V 1.380
37 R -0.557
38 V -0.724
39 C 0.568
40 V -0.739
41 K -0.510
42 K -0.514
43 S -0.966
44 T 0.399
45 Q -0.324
46 E -1.071
47 R -1.086
48 F -0.098
49 A 2.618
50 L 0.433
51 K 2.250
52 I -0.319
53 L 0.933
54 L -1.021
55 D -0.074
56 R -0.785
57 P -0.654
58 K -0.297
59 A -0.391
60 R -0.430
61 N -0.478
62 E 0.399
63 V -0.073
64 R -0.735
65 L -0.208
66 H 0.307
67 M -1.026
68 M -0.598
69 C -0.653
70 A -0.701
71 T -0.812
72 H -0.779
73 P -0.614
74 N -0.124
75 I 0.700
76 V 0.396
77 Q -0.874
78 I 0.308
79 I -0.788
80 E -0.211
81 V 0.104
82 F 0.327
83 A 0.140
84 N 0.957
85 S -0.718
86 V -1.000 *
87 Q -1.000 *
88 F -1.000 *
89 P -1.000 *
90 H -1.000 *
91 E -1.000 *
92 S -1.000 *
93 S -1.000 *
94 P -1.000 *
95 R -0.770
96 A -0.897
97 R -0.716
98 L 0.636
99 L 0.345
100 I 0.666
101 V 0.974
102 M 0.924
103 E 1.287
104 M -0.158
105 M 0.465
106 E -0.815
107 G 2.618
108 G 1.600
109 E 2.231
110 L 2.231
111 F 1.469
112 H -0.185
113 R 0.180
114 I 2.231
115 S -0.465
116 Q -0.520
117 H -0.489
118 R -0.204
119 H -0.796
120 F 0.991
121 T 0.844
122 E 1.897
123 K -0.356
124 Q -0.568
125 A 1.594
126 S 0.070
127 Q -1.114
128 V -0.123
129 T -0.488
130 K -0.459
131 Q -0.842
132 I 0.562
133 A -0.282
134 L -0.903
135 A 0.051
136 L 0.611
137 Q -1.009
138 H -0.938
139 C 0.452
140 H 1.839
141 L -1.053
142 L -0.480
143 N 0.141
144 I 1.388
145 A 0.046
146 H 2.231
147 R 2.231
148 D 2.618
149 L 1.428
150 K 2.618
151 P 2.618
152 E 2.231
153 N 2.618
154 L 0.948
155 L 1.727
156 F 0.926
157 K -0.758
158 D 0.039
159 N -0.899
160 S -0.797
161 L -0.680
162 D -0.579
163 A -0.201
164 P -0.965
165 V 0.972
166 K 0.751
167 L 0.122
168 C 0.490
169 D 1.678
170 F 1.842
171 G 2.618
172 F 0.369
173 A 1.485
174 K -0.576
175 V -0.668
176 D -0.698
177 Q -0.686
178 G -0.401
179 D -0.808
180 L -0.963
181 M -0.931
182 T -0.787
183 P -0.677
184 Q -0.194
185 F 1.292
186 T 1.439
187 P 0.318
188 Y 0.987
189 Y 2.155
190 V 0.578
191 A 1.850
192 P 2.258
193 Q 1.450
194 V 1.599
195 L 1.164
196 E -0.791
197 A 0.162
198 Q -0.401
199 R -1.000 *
200 R -1.000 *
201 H -1.000 *
202 Q -1.000 *
203 K -1.000 *
204 E -1.000 *
205 K -1.000 *
206 S -1.000 *
207 G -1.000 *
208 I -1.000 *
209 I -1.000 *
210 P -1.000 *
211 T -1.000 *
212 S -1.000 *
213 P -1.000 *
214 T -1.000 *
215 P -1.000 *
216 Y -1.000 *
217 T -1.000 *
218 Y 1.761
219 N 0.603
220 K -0.703
221 S 0.152
222 C 0.413
223 D 2.265
224 L 0.370
225 W 2.266
226 S 1.305
227 L 0.253
228 G 2.618
229 V 1.907
230 I 1.385
231 I 0.411
232 Y 1.668
233 V 1.804
234 M 1.236
235 L 1.521
236 C 0.542
237 G 2.618
238 Y -0.472
239 P 2.171
240 P 2.618
241 F 1.955
242 Y -0.398
243 S 0.274
244 K -0.303
245 H -0.438
246 H -0.345
247 S -1.000 *
248 R -1.000 *
249 T -1.000 *
250 I -1.000 *
251 P -1.000 *
252 K -0.597
253 D -0.351
254 M 0.858
255 R -0.086
256 K -0.623
257 K -0.243
258 I 1.706
259 M -0.383
260 T -1.007
261 G -0.018
262 S -1.085
263 F 0.108
264 E -0.262
265 F 1.039
266 P -0.722
267 E -0.836
268 E -0.435
269 E -0.129
270 W 1.675
271 S -0.654
272 Q -1.144
273 I 1.493
274 S 1.677
275 E -0.810
276 M -0.912
277 A 1.772
278 K 1.125
279 D -0.289
280 V 0.388
281 V 1.320
282 R -0.651
283 K -0.310
284 L 0.873
285 L 1.428
286 K -0.909
287 V -1.046
288 K -0.537
289 P 0.719
290 E -0.990
291 E -0.702
292 R 2.618
293 L -0.095
294 T 0.229
295 I 0.348
296 E -1.022
297 G 0.479
298 V 0.248
299 L 0.635
300 D -0.644
301 H 0.730
302 P -0.335
303 W 2.618
304 L 0.354
305 N -0.744
306 S -0.799
307 T -1.168
308 E -0.887
309 A -0.696
310 L -0.719
311 D -0.578
312 N -0.843
313 V -0.848
314 L -0.715
315 P -1.119
316 S -0.848
317 A -0.937
318 Q -0.660
319 L -0.656
320 M -0.974
321 M -0.892
322 D -0.778
323 K -0.916
324 A -0.834
325 V -0.879
326 V -0.876
327 A -0.666
328 G -0.831
329 I -0.456
330 Q -0.485
331 Q -0.710
332 A -0.299
333 H -0.553
334 A -0.321
335 E -0.788
336 Q -0.890
337 L 0.162
338 A -0.430
339 N -0.939
340 M -0.735
341 R -0.012
342 I -0.094
343 Q -1.105
344 D -1.034
345 L -0.619
346 K -0.641
347 V -1.053
348 S -0.934
349 L 0.450
350 K -0.874
351 P -0.927
352 L 0.017
353 H -0.370
354 S -0.822
355 V -0.588
356 N -0.592
357 N -0.032
358 P -0.278
359 I -0.970
360 L 0.216
361 R -0.563
362 K -0.719
363 R -0.629
364 K 0.212
365 L -0.534
366 L -0.733
367 G -0.934
368 T -0.526
369 K -0.875
370 P -1.020
371 K -0.879
372 D -1.124
373 G -0.181
374 I -0.877
375 Y -1.159
376 I -0.874
377 H -1.061
378 D -0.191
379 H -1.014
380 E -0.515
381 N -1.000 *
382 G -1.000 *
383 T -1.000 *
384 E -1.000 *
385 D -1.000 *
386 S -1.000 *
387 N -1.000 *
388 V -1.000 *
389 A -1.000 *
390 L -1.000 *
391 E -1.000 *
392 K -0.732
393 L -0.144
394 R -1.089
395 D -0.856
396 V -0.254
397 I -0.023
398 A -0.806
399 Q -0.799
400 C -0.636
401 I -0.390
402 L -0.691
403 P 0.113
404 Q -1.070
405 A -0.120
406 G 0.086
407 K -0.448
408 G -0.250
409 E -1.016
410 N -0.477
411 E -0.335
412 D -0.451
413 E -1.132
414 K 0.003
415 L 0.324
416 N -0.303
417 E -0.266
418 V 0.011
419 M -0.237
420 Q -0.588
421 E -0.552
422 A -0.655
423 W -1.016
424 K -0.459
425 Y -0.703
426 N -0.908
427 R -0.696
428 E -0.091
429 C -0.309
430 K -0.470
431 L -0.604
432 L -0.086
433 R -0.955
434 D -0.729
435 A 0.509
436 L 1.851
437 Q -0.685
438 S -0.706
439 F 0.900
440 S 1.169
441 W -0.524
442 N 0.777
443 G -0.507
444 R 0.027
445 G 0.083
446 F -0.412
447 T 0.250
448 D 0.255
449 K -0.834
450 V -1.000 *
451 D 0.580
452 R -0.281
453 L -0.890
454 K -0.264
455 L -0.942
456 A -0.893
457 E -0.763
458 V -1.018
459 V -0.750
460 K -0.649
461 Q -0.693
462 V -0.361
463 I -1.000 *
464 E -1.000 *
465 E -1.000 *
466 Q -1.000 *
467 T -1.000 *
468 L -1.000 *
469 P -1.000 *
470 H -1.000 *
471 E -1.000 *
472 P -1.000 *
473 Q -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 165.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MSEDSDMEKA IKETSILEEY SINWTQKLGA GISGPVRVCV KKSTQERFAL KILLDRPKAR
70 80 90 100 110 120
NEVRLHMMCA THPNIVQIIE VFANSVQFPH ESSPRARLLI VMEMMEGGEL FHRISQHRHF
130 140 150 160 170 180
TEKQASQVTK QIALALQHCH LLNIAHRDLK PENLLFKDNS LDAPVKLCDF GFAKVDQGDL
190 200 210 220 230 240
MTPQFTPYYV APQVLEAQRR HQKEKSGIIP TSPTPYTYNK SCDLWSLGVI IYVMLCGYPP
250 260 270 280 290 300
FYSKHHSRTI PKDMRKKIMT GSFEFPEEEW SQISEMAKDV VRKLLKVKPE ERLTIEGVLD
310 320 330 340 350 360
HPWLNSTEAL DNVLPSAQLM MDKAVVAGIQ QAHAEQLANM RIQDLKVSLK PLHSVNNPIL
370 380 390 400 410 420
RKRKLLGTKP KDGIYIHDHE NGTEDSNVAL EKLRDVIAQC ILPQAGKGEN EDEKLNEVMQ
430 440 450 460 470
EAWKYNRECK LLRDALQSFS WNGRGFTDKV DRLKLAEVVK QVIEEQTLPH EPQ
2ONL (X-Ray,4.00 Å resolution)
2OZA (X-Ray,2.70 Å resolution)
Although activation of the cAMP/PKA pathway by the cAMP-elevating agent forskolin has been shown to trigger
a transient nuclear export of MK5, the S115D mutant resided predominantly in the nucleus in forskolin-treated cells. PKA-mediated phosphorylation of MK5 at Ser-115 is implicated in nuclear export of MK5. Furthermore, the nuclear expression of a double EGFP-MK5 S115D/L337A mutant suggested that a functional NES is required for PKA-mediated nuclear exclusion of MK5.
ERK3-,ERK4-, and p38-MAPK-mediated nuclear export of MK5 does not require Serine115. PKI-like and Rev-like NES were identified, but not experimentally proven to be sufficient.
An earlier paper from the same group showed that stress-induced nuclear export of MK5 also relies on CRM1 pathway and the signal is likely located from 345-352.
Doubts about NES: MK5-(337-355) failed to bind CRM1 in GST-pulldown assay (Chook Lab, unpublished results).
[1]. "Serine residue 115 of MAPK-activated protein kinase MK5 is crucial for its PKA-regulated nuclear export and biological function."
Kostenko S, Shiryaev A, Gerits N, Dumitriu G, Klenow H, Johannessen M, Moens U. (2010)
Cell Mol Life Sci,
:
PubMed[2]. "Both binding and activation of p38 mitogen-activated protein kinase (MAPK) play essential roles in regulation of the nucleocytoplasmic distribution of MAPK-activated protein kinase 5 by cellular stress"
Seternes OM, Johansen B, Hegge B, Johannessen M, Keyse SM, Moens U. (2002)
Mol Cell Biol,
22:6931-45
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
