Pre-B-cell leukemia transcription factor 1
UniProt
Show FASTA Format
>gi|20141754|sp|P41778.2|PBX1_MOUSE RecName: Full=Pre-B-cell leukemia transcription factor 1; AltName: Full=Homeobox protein PBX1
MDEQPRLMHSHAGVGMAGHPGLSQHLQDGAGGTEGEGGRKQDIGDILQQIMTITDQSLDEAQARKHALNC
HRMKPALFNVLCEIKEKTVLSIRGAQEEEPTDPQLMRLDNMLLAEGVAGPEKGGGSAAAAAAAAASGGAG
SDNSVEHSDYRAKLSQIRQIYHTELEKYEQACNEFTTHVMNLLREQSRTRPISPKEIERMVSIIHRKFSS
IQMQLKQSTCEAVMILRSRFLDARRKRRNFNKQATEILNEYFYSHLSNPYPSEEAKEELAKKCGITVSQV
SNWFGNKRIRYKKNIGKFQEEANIYAAKTAVTATNVSAHGSQANSPSTPNSAGSSSSFNMSNSGDLFMSV
QSLNGDSYQGAQVGANVQSQVDTLRHVISQTGGYSDGLAASQMYSPQGISANGGWQDATTPSSVTSPTEG
PGSVHSDTSN
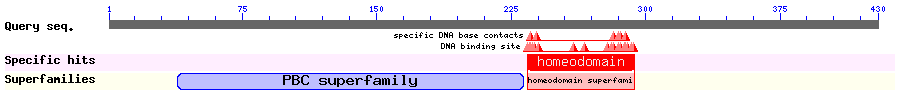
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 986321112489998999999998998999999999998788489887663035679249
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHCCCCCCHHH
AA: MDEQPRLMHSHAGVGMAGHPGLSQHLQDGAGGTEGEGGRKQDIGDILQQIMTITDQSLDE
10 20 30 40 50 60
Conf: 997211003577760367889987765421014788889998576423435430654489
Pred: HHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHCCCCCC
AA: AQARKHALNCHRMKPALFNVLCEIKEKTVLSIRGAQEEEPTDPQLMRLDNMLLAEGVAGP
70 80 90 100 110 120
Conf: 999997146667863399999998661369999999999999999999998755667899
Pred: CCCCCCHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH
AA: EKGGGSAAAAAAAAASGGAGSDNSVEHSDYRAKLSQIRQIYHTELEKYEQACNEFTTHVM
130 140 150 160 170 180
Conf: 997540267889978873243201455214678765668789987630010112210245
Pred: HHHHHHCCCCCCCHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHCCCHHHHHHCCC
AA: NLLREQSRTRPISPKEIERMVSIIHRKFSSIQMQLKQSTCEAVMILRSRFLDARRKRRNF
190 200 210 220 230 240
Conf: 146899999998842169999968999999884741641012101331221344035778
Pred: CHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHCCEEEEECCCCCCCCCCCCCCCHHHHH
AA: NKQATEILNEYFYSHLSNPYPSEEAKEELAKKCGITVSQVSNWFGNKRIRYKKNIGKFQE
250 260 270 280 290 300
Conf: 866754201244454567999999999999999999988899877545555788999989
Pred: HHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: EANIYAAKTAVTATNVSAHGSQANSPSTPNSAGSSSSFNMSNSGDLFMSVQSLNGDSYQG
310 320 330 340 350 360
Conf: 867664334454444445688999998787778998754689999999999998999999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: AQVGANVQSQVDTLRHVISQTGGYSDGLAASQMYSPQGISANGGWQDATTPSSVTSPTEG
370 380 390 400 410 420
Conf: 9998989999
Pred: CCCCCCCCCC
AA: PGSVHSDTSN
430
Show Conservation Score by AL2CO
1 M -1.000 *
2 D -1.000 *
3 E -1.000 *
4 Q -1.000 *
5 P -1.000 *
6 R -1.000 *
7 L -1.000 *
8 M -1.000 *
9 H -1.000 *
10 S -1.000 *
11 H -1.000 *
12 A -1.000 *
13 G -1.000 *
14 V -1.000 *
15 G -1.000 *
16 M -1.000 *
17 A -1.000 *
18 G -1.000 *
19 H -1.000 *
20 P -1.000 *
21 G -1.000 *
22 L -1.000 *
23 S -1.000 *
24 Q -1.000 *
25 H -1.000 *
26 L -1.000 *
27 Q -1.000 *
28 D -1.000 *
29 G -1.000 *
30 A -1.000 *
31 G -1.000 *
32 G -1.000 *
33 T -1.000 *
34 E -1.000 *
35 G -1.000 *
36 E -1.000 *
37 G -1.000 *
38 G -1.000 *
39 R -1.000 *
40 K -1.000 *
41 Q -1.000 *
42 D -1.000 *
43 I -1.000 *
44 G -1.000 *
45 D -1.000 *
46 I -1.000 *
47 L -1.000 *
48 Q -1.000 *
49 Q -1.000 *
50 I -1.000 *
51 M -1.000 *
52 T -1.000 *
53 I -1.000 *
54 T -1.000 *
55 D -1.000 *
56 Q -1.000 *
57 S -1.000 *
58 L -1.000 *
59 D -1.000 *
60 E -1.000 *
61 A -1.000 *
62 Q -1.000 *
63 A -1.000 *
64 R -1.000 *
65 K -1.000 *
66 H -1.000 *
67 A -1.000 *
68 L -1.000 *
69 N -1.000 *
70 C -1.000 *
71 H -1.000 *
72 R -1.000 *
73 M -1.000 *
74 K -1.000 *
75 P -1.000 *
76 A -1.000 *
77 L -1.000 *
78 F -1.000 *
79 N -1.000 *
80 V -1.000 *
81 L -1.000 *
82 C -1.000 *
83 E -1.000 *
84 I -1.000 *
85 K -1.000 *
86 E -1.000 *
87 K -1.000 *
88 T -1.000 *
89 V -0.516
90 L -0.447
91 S -0.409
92 I -0.306
93 R 0.434
94 G -0.315
95 A -0.594
96 Q -0.177
97 E -0.214
98 E 0.050
99 E -0.384
100 P 0.140
101 T -0.349
102 D -0.277
103 P -0.057
104 Q -0.019
105 L 0.111
106 M -0.243
107 R 0.133
108 L -0.145
109 D 0.075
110 N -0.368
111 M -0.327
112 L -0.540
113 L -0.512
114 A 0.049
115 E 0.217
116 G -0.153
117 V 0.036
118 A -0.335
119 G -0.322
120 P -0.207
121 E -0.397
122 K -0.177
123 G -0.567
124 G -0.146
125 G -0.594
126 S -0.388
127 A -0.564
128 A -0.620
129 A -0.424
130 A -0.614
131 A -0.158
132 A -0.790
133 A -0.418
134 A -0.681
135 A -0.696
136 S -0.025
137 G -0.507
138 G -0.633
139 A -0.837
140 G -0.569
141 S -0.860
142 D -0.502
143 N -0.342
144 S -0.385
145 V -0.597
146 E 0.227
147 H -0.491
148 S -0.568
149 D 0.692
150 Y -0.200
151 R 0.174
152 A -0.633
153 K 0.620
154 L 0.757
155 S -0.272
156 Q 0.738
157 I 0.539
158 R 0.282
159 Q -0.317
160 I -0.278
161 Y 0.760
162 H -0.403
163 T -0.222
164 E 0.881
165 L 0.169
166 E 0.095
167 K 0.518
168 Y 0.741
169 E 0.046
170 Q 0.445
171 A -0.108
172 C 0.238
173 N -0.348
174 E -0.090
175 F 0.485
176 T 0.109
177 T -0.316
178 H -0.147
179 V 0.460
180 M -0.433
181 N 0.117
182 L -0.146
183 L 0.737
184 R -0.125
185 E -0.242
186 Q -0.498
187 S 0.019
188 R -0.394
189 T -0.242
190 R -0.456
191 P 0.697
192 I -0.202
193 S 0.136
194 P -0.374
195 K -0.397
196 E 0.192
197 I -0.286
198 E -0.369
199 R -0.270
200 M -0.573
201 V -0.542
202 S -0.394
203 I -0.833
204 I -0.110
205 H -0.232
206 R -0.518
207 K -0.442
208 F -0.646
209 S -0.329
210 S -0.751
211 I -0.572
212 Q -0.531
213 M -0.661
214 Q -0.554
215 L -0.392
216 K -0.034
217 Q -0.526
218 S -0.393
219 T -0.659
220 C -0.576
221 E -0.834
222 A -0.504
223 V -0.602
224 M -0.775
225 I -0.456
226 L -0.125
227 R -0.538
228 S -0.222
229 R -0.290
230 F -0.547
231 L -0.730
232 D 0.080
233 A -0.479
234 R -0.474
235 R 0.795
236 K 1.356
237 R 0.534
238 R 1.096
239 N 0.075
240 F 1.604
241 N 0.569
242 K 0.907
243 Q -0.247
244 A 1.547
245 T 0.795
246 E 0.135
247 I 0.839
248 L 2.257
249 N 0.967
250 E 0.422
251 Y 2.188
252 F 1.233
253 Y 0.435
254 S 0.498
255 H 3.469
256 L 0.326
257 S -0.399
258 N 2.096
259 P 3.616
260 Y 4.557
261 P 4.007
262 S 1.757
263 E 1.855
264 E 0.345
265 A 0.928
266 K 3.111
267 E -0.159
268 E -0.155
269 L 3.074
270 A 1.656
271 K 0.180
272 K 0.402
273 C 1.437
274 G 0.664
275 I 2.606
276 T 1.207
277 V 0.412
278 S 0.340
279 Q 4.557
280 V 2.753
281 S 1.154
282 N 2.880
283 W 3.044
284 F 2.637
285 G 1.412
286 N 3.082
287 K 1.352
288 R 2.608
289 I 0.971
290 R 2.760
291 Y 0.069
292 K 1.106
293 K 0.782
294 N 0.114
295 I 0.560
296 G 0.015
297 K 0.158
298 F -0.506
299 Q -0.176
300 E -0.239
301 E -0.291
302 A -0.572
303 N -0.546
304 I -0.637
305 Y -0.751
306 A -0.537
307 A -0.618
308 K -0.476
309 T -0.699
310 A -0.352
311 V -0.901
312 T -0.758
313 A -0.501
314 T -0.689
315 N -0.788
316 V -0.937
317 S -0.711
318 A -0.770
319 H -0.759
320 G -0.681
321 S -0.741
322 Q -0.736
323 A -0.800
324 N -0.624
325 S -0.783
326 P -0.817
327 S -0.718
328 T -0.475
329 P -0.596
330 N -0.680
331 S -0.541
332 A -0.817
333 G -0.873
334 S -0.944
335 S -0.905
336 S -0.812
337 S -0.675
338 F -0.971
339 N -0.717
340 M -0.729
341 S -0.787
342 N -0.870
343 S -0.675
344 G -0.619
345 D -0.560
346 L -0.911
347 F -0.884
348 M -0.836
349 S -0.719
350 V -0.642
351 Q -0.782
352 S -0.726
353 L -0.658
354 N -0.695
355 G -0.782
356 D -0.752
357 S -0.556
358 Y -0.791
359 Q -0.754
360 G -0.703
361 A -0.847
362 Q -0.739
363 V -0.571
364 G -0.655
365 A -0.832
366 N -0.755
367 V -0.794
368 Q -0.718
369 S -0.623
370 Q -0.661
371 V -0.695
372 D -0.549
373 T -0.787
374 L -0.590
375 R -0.799
376 H -1.000 *
377 V -1.000 *
378 I -1.000 *
379 S -1.000 *
380 Q -1.000 *
381 T -1.000 *
382 G -1.000 *
383 G -1.000 *
384 Y -1.000 *
385 S -1.000 *
386 D -1.000 *
387 G -1.000 *
388 L -1.000 *
389 A -1.000 *
390 A -1.000 *
391 S -1.000 *
392 Q -1.000 *
393 M -1.000 *
394 Y -1.000 *
395 S -1.000 *
396 P -1.000 *
397 Q -1.000 *
398 G -1.000 *
399 I -1.000 *
400 S -1.000 *
401 A -1.000 *
402 N -1.000 *
403 G -1.000 *
404 G -1.000 *
405 W -1.000 *
406 Q -1.000 *
407 D -1.000 *
408 A -1.000 *
409 T -1.000 *
410 T -1.000 *
411 P -1.000 *
412 S -1.000 *
413 S -1.000 *
414 V -1.000 *
415 T -1.000 *
416 S -1.000 *
417 P -1.000 *
418 T -1.000 *
419 E -1.000 *
420 G -1.000 *
421 P -1.000 *
422 G -1.000 *
423 S -1.000 *
424 V -1.000 *
425 H -1.000 *
426 S -1.000 *
427 D -1.000 *
428 T -1.000 *
429 S -1.000 *
430 N -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 172.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MDEQPRLMHS HAGVGMAGHP GLSQHLQDGA GGTEGEGGRK QDIGDILQQI MTITDQSLDE
70 80 90 100 110 120
AQARKHALNC HRMKPALFNV LCEIKEKTVL SIRGAQEEEP TDPQLMRLDN MLLAEGVAGP
130 140 150 160 170 180
EKGGGSAAAA AAAAASGGAG SDNSVEHSDY RAKLSQIRQI YHTELEKYEQ ACNEFTTHVM
190 200 210 220 230 240
NLLREQSRTR PISPKEIERM VSIIHRKFSS IQMQLKQSTC EAVMILRSRF LDARRKRRNF
250 260 270 280 290 300
NKQATEILNE YFYSHLSNPY PSEEAKEELA KKCGITVSQV SNWFGNKRIR YKKNIGKFQE
310 320 330 340 350 360
EANIYAAKTA VTATNVSAHG SQANSPSTPN SAGSSSSFNM SNSGDLFMSV QSLNGDSYQG
370 380 390 400 410 420
AQVGANVQSQ VDTLRHVISQ TGGYSDGLAA SQMYSPQGIS ANGGWQDATT PSSVTSPTEG
430
PGSVHSDTSN
1B8I (X-Ray,2.40 Å resolution)
PBX1 is a member of the PBC subfamily of TALE homodomain proteins. Berthelsen eta al. showed that the PBC-A domain of PBX1 is required for the export of PBX1 in a LMB dependent manner. Kilstrup-Nielsen et al. further characterized the NES by demonstrating that aa 73-90 is sufficient for CRM1 export and the PBC-A domain can bind CRM1. However, they found that PBXΔ73-90 can still be exported, while PBXΔ45-90 cannot. Nevertheless, aa 45-72 is not sufficient to export GFP. It seems that aa 45-52 also plays a role in export. No point mutations were made to pinpoint critical residues.
Ref.1The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH., Berthelsen et al., Genes Dev, 1999 Ref.2PBX1 nuclear export is regulated independently of PBX-MEINOX interaction by PKA phosphorylation of the PBC-B domain, Kilstrup-Nielsen et al., EMBO J, 2003
[1]. "The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH."
Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. (1999)
Genes Dev,
13:946-53
PubMed[2]. "PBX1 nuclear export is regulated independently of PBX-MEINOX interaction by PKA phosphorylation of the PBC-B domain"
Kilstrup-Nielsen C, Alessio M, Zappavigna V. (2003)
EMBO J,
22:89-99
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
