Tyrosine-protein kinase Fyn
UniProt
Show FASTA Format
>gi|143811392|sp|P39688.3|FYN_MOUSE RecName: Full=Tyrosine-protein kinase Fyn; AltName: Full=Proto-oncogene c-Fyn; AltName: Full=p59-Fyn
MGCVQCKDKEAAKLTEERDGSLNQSSGYRYGTDPTPQHYPSFGVTSIPNYNNFHAAGGQGLTVFGGVNSS
SHTGTLRTRGGTGVTLFVALYDYEARTEDDLSFHKGEKFQILNSSEGDWWEARSLTTGETGYIPSNYVAP
VDSIQAEEWYFGKLGRKDAERQLLSFGNPRGTFLIRESQTTKGAYSLSIRDWDDMKGDHVKHYKIRKLDN
GGYYITTRAQFETLQQLVQHYSERAAGLCCRLVVPCHKGMPRLTDLSVKTKDVWEIPRESLQLIKRLGNG
QFGEVWLGTWNGNTKVAIKTLKPGTMSPESFLEEAQIMKKLKHDKLVQLYAVVSEEPIYIVTEYMSKGSL
LDFLKDGEGRALKLPNLVDMAAQVAAGMAYIERMNYIHRDLRSANILVGNGLICKIADFGLARLIEDNEY
TARQGAKFPIKWTAPEAALYGRFTIKSDVWSFGILLTELVTKGRVPYPGMNNREVLEQVERGYRMPCPQD
CPISLHELMIHCWKKDPEERPTFEYLQGFLEDYFTATEPQYQPGENL
Show Domain Info by CDD
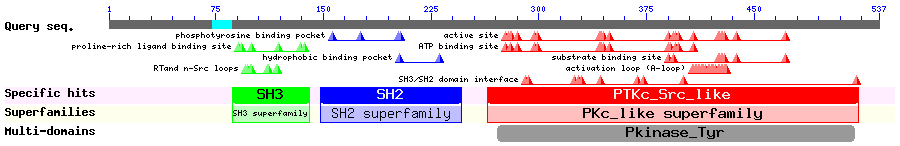
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 986445553210000012799899999866899999999998855789999988789997
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: MGCVQCKDKEAAKLTEERDGSLNQSSGYRYGTDPTPQHYPSFGVTSIPNYNNFHAAGGQG
10 20 30 40 50 60
Conf: 200277777776676555699872799994117778878876456653798107987601
Pred: CCCCCCCCCCCCCCCCCCCCCCCCEEEEEEECCCCCCCCCCCCCCCCEEEEEECCCCCCE
AA: LTVFGGVNSSSHTGTLRTRGGTGVTLFVALYDYEARTEDDLSFHKGEKFQILNSSEGDWW
70 80 90 100 110 120
Conf: 100045687345468850028985323211375770037764024899850168740678
Pred: ECCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHCCCCCCCEEEEEEECCC
AA: EARSLTTGETGYIPSNYVAPVDSIQAEEWYFGKLGRKDAERQLLSFGNPRGTFLIRESQT
130 140 150 160 170 180
Conf: 886136664127788998011013344079873584153302199999985205764310
Pred: CCCEEEEEEECCCCCCCCCEEEEEEEECCCCCEEEECCCCHHHHHHHHHHHHHCCCCCCC
AA: TKGAYSLSIRDWDDMKGDHVKHYKIRKLDNGGYYITTRAQFETLQQLVQHYSERAAGLCC
190 200 210 220 230 240
Conf: 025434679987568987877532213301467767159754512244315831799722
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHCCCCCCCEEEEEECCCEEEEEEC
AA: RLVVPCHKGMPRLTDLSVKTKDVWEIPRESLQLIKRLGNGQFGEVWLGTWNGNTKVAIKT
250 260 270 280 290 300
Conf: 589879978689999887640523505234541258327887513677431011367788
Pred: CCCCCCCHHHHHHHHHHHHHHCCCCEEEEEEEECCCCEEEEEEECCCCCCCCCCCCCCCC
AA: LKPGTMSPESFLEEAQIMKKLKHDKLVQLYAVVSEEPIYIVTEYMSKGSLLDFLKDGEGR
310 320 330 340 350 360
Conf: 558730442389877333678773101223220131002641134202430010015653
Pred: CCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCEECCCEEEEECCCCCEEECCCCCC
AA: ALKLPNLVDMAAQVAAGMAYIERMNYIHRDLRSANILVGNGLICKIADFGLARLIEDNEY
370 380 390 400 410 420
Conf: 322688776434611221023217812310145565555304987899999489999756
Pred: CCCCCCCCCCCCCCHHHHCCCCEEEECCCCHHHHHHHHHHHCCCCCCCCCCCHHHHHHHH
AA: TARQGAKFPIKWTAPEAALYGRFTIKSDVWSFGILLTELVTKGRVPYPGMNNREVLEQVE
430 440 450 460 470 480
Conf: 124899999990669999998613599999706787876543221579899999999
Pred: HCCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCCCC
AA: RGYRMPCPQDCPISLHELMIHCWKKDPEERPTFEYLQGFLEDYFTATEPQYQPGENL
490 500 510 520 530
Show Conservation Score by AL2CO
1 M -1.000 *
2 G -1.000 *
3 C -1.000 *
4 V -1.000 *
5 Q -1.000 *
6 C -1.000 *
7 K -1.000 *
8 D -1.000 *
9 K -1.000 *
10 E -1.000 *
11 A -1.000 *
12 A -1.000 *
13 K -1.000 *
14 L -1.000 *
15 T -1.000 *
16 E -1.000 *
17 E -1.000 *
18 R -1.000 *
19 D -1.000 *
20 G -1.000 *
21 S -1.000 *
22 L -1.000 *
23 N -1.000 *
24 Q -1.000 *
25 S -1.000 *
26 S -1.000 *
27 G -1.000 *
28 Y -1.000 *
29 R -1.000 *
30 Y -1.000 *
31 G -1.000 *
32 T -1.000 *
33 D -1.000 *
34 P -1.000 *
35 T -1.000 *
36 P -1.000 *
37 Q -1.000 *
38 H -1.000 *
39 Y -1.000 *
40 P -1.000 *
41 S -1.000 *
42 F -1.000 *
43 G -1.000 *
44 V -1.000 *
45 T -1.000 *
46 S -1.000 *
47 I -1.000 *
48 P -1.000 *
49 N -1.000 *
50 Y -1.000 *
51 N -1.000 *
52 N -1.000 *
53 F -1.000 *
54 H -1.000 *
55 A -1.025
56 A -1.075
57 G -1.070
58 G -1.078
59 Q -0.903
60 G -0.989
61 L -1.127
62 T -1.112
63 V -1.077
64 F -1.097
65 G -1.084
66 G -0.993
67 V -1.054
68 N -1.121
69 S -1.062
70 S -1.131
71 S -1.087
72 H -1.032
73 T -1.016
74 G -1.050
75 T -0.952
76 L -1.141
77 R -1.054
78 T -1.151
79 R -1.082
80 G -1.053
81 G -1.068
82 T -1.024
83 G -0.896
84 V -1.064
85 T -1.098
86 L -0.908
87 F -0.008
88 V -0.490
89 A 0.750
90 L -0.002
91 Y 0.263
92 D -0.190
93 Y 0.785
94 E -0.878
95 A -0.010
96 R -0.702
97 T -0.744
98 E -0.787
99 D -0.835
100 D 0.298
101 L 0.814
102 S -0.601
103 F 0.594
104 H -0.824
105 K -0.208
106 G 0.017
107 E 0.248
108 K -0.711
109 F 0.503
110 Q -0.998
111 I 0.142
112 L -0.426
113 N -0.623
114 S -0.842
115 S -0.980
116 E -0.657
117 G -0.919
118 D -0.628
119 W 0.482
120 W -0.206
121 E -0.886
122 A 0.066
123 R -0.658
124 S -0.576
125 L -0.875
126 T -0.913
127 T -0.981
128 G -0.575
129 E -0.983
130 T -0.881
131 G 0.430
132 Y -0.029
133 I 0.710
134 P 2.182
135 S -0.870
136 N 0.104
137 Y 0.841
138 V 0.960
139 A -0.557
140 P -0.670
141 V -1.049
142 D -0.707
143 S -0.625
144 I -0.280
145 Q -0.625
146 A -0.891
147 E -0.437
148 E -0.665
149 W 0.691
150 Y 0.801
151 F -0.594
152 G -0.529
153 K -0.833
154 L 0.302
155 G -0.234
156 R 1.123
157 K -0.797
158 D -0.265
159 A 0.784
160 E -0.642
161 R -0.640
162 Q -0.855
163 L 1.290
164 L -0.500
165 S -1.030
166 F -1.059
167 G -0.963
168 N -0.818
169 P -0.964
170 R -0.891
171 G 1.236
172 T -0.246
173 F 1.394
174 L 0.548
175 I 0.763
176 R 1.921
177 E -0.703
178 S 0.428
179 Q -0.290
180 T -0.221
181 T -0.910
182 K -0.966
183 G -0.456
184 A -0.785
185 Y 0.204
186 S 0.178
187 L 0.372
188 S 0.399
189 I 0.033
190 R -0.650
191 D -0.613
192 W -1.170
193 D -0.772
194 D -0.759
195 M -1.109
196 K -1.003
197 G -0.708
198 D -0.936
199 H -0.912
200 V 0.023
201 K -0.270
202 H 1.111
203 Y 1.162
204 K 0.058
205 I 1.133
206 R -0.655
207 K -0.954
208 L -0.915
209 D -0.726
210 N -0.859
211 G -0.431
212 G -0.728
213 Y 0.318
214 Y 0.231
215 I 0.648
216 T -0.128
217 T -0.661
218 R -0.692
219 A -0.955
220 Q -1.031
221 F 0.544
222 E -0.842
223 T -0.103
224 L 0.897
225 Q -1.079
226 Q -0.462
227 L 0.838
228 V 1.027
229 Q -1.033
230 H 0.482
231 Y 1.383
232 S -0.699
233 E -0.910
234 R -0.819
235 A -0.820
236 A -0.410
237 G -0.152
238 L 0.004
239 C -0.639
240 C -0.809
241 R -0.857
242 L 0.563
243 V -0.964
244 V -1.074
245 P 0.125
246 C -0.400
247 H -1.096
248 K -1.010
249 G -1.178
250 M -1.032
251 P -0.948
252 R -1.051
253 L -0.713
254 T -0.981
255 D -0.921
256 L -0.793
257 S -0.835
258 V -0.970
259 K -0.918
260 T -1.045
261 K -1.103
262 D 0.105
263 V -0.757
264 W 0.767
265 E 0.875
266 I 0.029
267 P -0.260
268 R 0.466
269 E -0.741
270 S -0.264
271 L 0.865
272 Q -0.802
273 L 0.205
274 I -0.963
275 K -0.513
276 R -0.201
277 L 0.704
278 G 2.926
279 N -0.522
280 G 2.118
281 Q -0.403
282 F 1.378
283 G 1.232
284 E -0.102
285 V 1.119
286 W -0.123
287 L -0.646
288 G 0.949
289 T -0.805
290 W -0.038
291 N -0.208
292 G -0.632
293 N -1.006
294 T -0.931
295 K -0.582
296 V 1.212
297 A 2.094
298 I 1.459
299 K 2.006
300 T -0.217
301 L 0.839
302 K 0.176
303 P -0.724
304 G 0.238
305 T -0.437
306 M 0.374
307 S -0.785
308 P -0.667
309 E -0.716
310 S -0.293
311 F 1.073
312 L -0.639
313 E -0.465
314 E 2.067
315 A 0.480
316 Q -0.425
317 I -0.016
318 M 1.507
319 K 0.657
320 K -0.479
321 L 0.406
322 K -0.166
323 H 2.100
324 D -0.647
325 K 0.977
326 L 2.000
327 V 1.003
328 Q -0.186
329 L 2.443
330 Y -0.249
331 A 1.426
332 V 0.704
333 V 0.746
334 S 0.187
335 E -0.450
336 E -0.502
337 P 1.313
338 I 0.229
339 Y 0.350
340 I 0.648
341 V 0.957
342 T 0.022
343 E 1.661
344 Y 0.513
345 M 0.901
346 S -0.899
347 K -0.479
348 G 1.459
349 S 0.857
350 L 2.430
351 L -0.100
352 D -0.526
353 F 0.842
354 L 1.490
355 K -0.172
356 D -0.640
357 G -0.896
358 E -0.791
359 G -0.717
360 R -0.969
361 A -1.125
362 L -0.699
363 K -0.946
364 L -0.867
365 P -1.094
366 N -1.141
367 L 0.263
368 V 0.422
369 D -0.605
370 M 0.312
371 A 0.103
372 A -0.771
373 Q 0.295
374 V 0.936
375 A 0.319
376 A -0.558
377 G 1.361
378 M 1.897
379 A -0.725
380 Y 0.654
381 I 1.240
382 E 2.001
383 R -0.767
384 M -0.493
385 N -0.403
386 Y 0.013
387 I 1.047
388 H 2.434
389 R 1.554
390 D 2.001
391 L 1.713
392 R 1.246
393 S 0.869
394 A 1.242
395 N 2.001
396 I 0.673
397 L 2.434
398 V 0.647
399 G -0.054
400 N -0.602
401 G -1.010
402 L -0.805
403 I -0.670
404 C 0.464
405 K 1.846
406 I 1.348
407 A 0.604
408 D 1.949
409 F 1.569
410 G 1.949
411 L 1.644
412 A 0.939
413 R 1.207
414 L -0.195
415 I 0.090
416 E -0.813
417 D 0.116
418 N -0.322
419 E -0.705
420 Y 0.482
421 T -0.755
422 A -0.168
423 R -0.634
424 Q -1.003
425 G -0.534
426 A -0.393
427 K 0.335
428 F 0.703
429 P 1.096
430 I 0.646
431 K 1.652
432 W 2.367
433 T 0.677
434 A 1.176
435 P 2.171
436 E 1.871
437 A 1.106
438 A 0.084
439 L -0.848
440 Y -0.806
441 G -0.472
442 R -0.586
443 F 1.951
444 T 1.719
445 I 0.111
446 K 2.337
447 S 2.154
448 D 2.926
449 V 1.280
450 W 2.355
451 S 1.958
452 F 1.349
453 G 1.897
454 I 1.698
455 L 0.719
456 L 0.706
457 T -0.084
458 E 2.392
459 L 0.877
460 V -0.134
461 T 0.993
462 K -0.527
463 G 2.419
464 R -0.241
465 V -0.610
466 P 2.403
467 Y 1.665
468 P -0.238
469 G -0.360
470 M -0.605
471 N -0.700
472 N 0.051
473 R -0.890
474 E 0.039
475 V 0.759
476 L -0.135
477 E -0.713
478 Q -0.876
479 V 1.209
480 E -0.688
481 R -0.832
482 G 0.275
483 Y -0.275
484 R 1.262
485 M 1.390
486 P -0.554
487 C -0.381
488 P 0.713
489 Q -1.046
490 D -1.029
491 C 0.332
492 P -0.334
493 I -0.896
494 S -1.048
495 L 0.227
496 H 1.226
497 E -0.761
498 L 0.321
499 M 0.933
500 I -1.030
501 H -0.979
502 C 0.697
503 W 2.926
504 K -0.706
505 K -0.984
506 D -0.896
507 P 0.181
508 E -0.751
509 E -0.611
510 R 2.926
511 P 1.926
512 T -0.433
513 F 2.926
514 E -0.803
515 Y -0.958
516 L 0.903
517 Q -0.805
518 G -1.038
519 F -0.986
520 L 0.984
521 E -0.713
522 D -0.492
523 Y -0.101
524 F -0.663
525 T -0.747
526 A -0.784
527 T -0.273
528 E -0.709
529 P -0.854
530 Q -0.477
531 Y 1.069
532 Q -0.506
533 P -1.000 *
534 G -1.000 *
535 E -1.000 *
536 N -1.000 *
537 L -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 188.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MGCVQCKDKE AAKLTEERDG SLNQSSGYRY GTDPTPQHYP SFGVTSIPNY NNFHAAGGQG
70 80 90 100 110 120
LTVFGGVNSS SHTGTLRTRG GTGVTLFVAL YDYEARTEDD LSFHKGEKFQ ILNSSEGDWW
130 140 150 160 170 180
EARSLTTGET GYIPSNYVAP VDSIQAEEWY FGKLGRKDAE RQLLSFGNPR GTFLIRESQT
190 200 210 220 230 240
TKGAYSLSIR DWDDMKGDHV KHYKIRKLDN GGYYITTRAQ FETLQQLVQH YSERAAGLCC
250 260 270 280 290 300
RLVVPCHKGM PRLTDLSVKT KDVWEIPRES LQLIKRLGNG QFGEVWLGTW NGNTKVAIKT
310 320 330 340 350 360
LKPGTMSPES FLEEAQIMKK LKHDKLVQLY AVVSEEPIYI VTEYMSKGSL LDFLKDGEGR
370 380 390 400 410 420
ALKLPNLVDM AAQVAAGMAY IERMNYIHRD LRSANILVGN GLICKIADFG LARLIEDNEY
430 440 450 460 470 480
TARQGAKFPI KWTAPEAALY GRFTIKSDVW SFGILLTELV TKGRVPYPGM NNREVLEQVE
490 500 510 520 530
RGYRMPCPQD CPISLHELMI HCWKKDPEER PTFEYLQGFL EDYFTATEPQ YQPGENL
Fyn is an SRC kinase family protein involved in functions including T-cell signaling, mitogenic signaling, and cell adhesion-mediated signaling. Fyn is exported after exposure to oxidative stress, and is ubiquitinated and degraded upon nuclear export. Fyn export is also controlled by tyrosine phosphorylation of residue 213 as well as the leucine-rich NES.
Fyn is a negative regulator of Nrf2. The nuclear export of Nrf2 is activated as Fyn accumulates inside the nucleus and phosphorylates tyrosine 586 of Nrf2, which leads to CRM1-mediated nuclear export of Nrf2.
[1]. "Tyrosine phosphorylation controls nuclear export of Fyn, allowing Nrf2 activation of cytoprotective gene expression."
Kaspar JW, Jaiswal AK. (2010)
FASEB J,
:
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
