Summary for Cdk5-p27 (NES ID: 194)
Full Name
Cell division protein kinase 5
UniProt Alternative Names
Cyclin-dependent kinase 5, Serine/threonine-protein kinase PSSALRE, Tau protein kinase II catalytic subunit
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
Secondary Structure of Export Signal
β-sheet (residues 70-79)
β-strands and loop (residues 128-142)
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|4033704|sp|Q00535.3|CDK5_HUMAN RecName: Full=Cell division protein kinase 5; AltName: Full=Cyclin-dependent kinase 5; AltName: Full=Serine/threonine-protein kinase PSSALRE; AltName: Full=Tau protein kinase II catalytic subunit; Short=TPKII catalytic subunit
MQKYEKLEKIGEGTYGTVFKAKNRETHEIVALKRVRLDDDDEGVPSSALREICLLKELKHKNIVRLHDVL
HSDKKLTLVFEFCDQDLKKYFDSCNGDLDPEIVKSFLFQLLKGLGFCHSRNVLHRDLKPQNLLINRNGEL
KLADFGLARAFGIPVRCYSAEVVTLWYRPPDVLFGAKLYSTSIDMWSAGCIFAELANAGRPLFPGNDVDD
QLKRIFRLLGTPTEEQWPSMTKLPDYKPYPMYPATTSLVNVVPKLNATGRDLLQNLLKCNPVQRISAEEA
LQHPYFSDFCPP
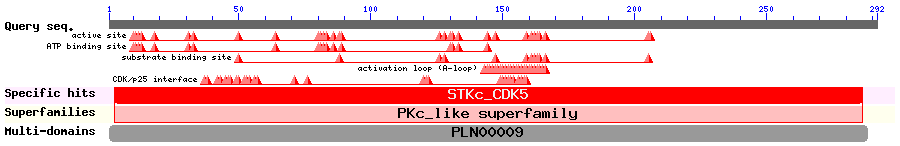
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 900024332246411577643037888358863101588999998246888663201799
Pred: CCHHHHHHHCCCCCEEEEEEEECCCCCCEEEEEECCCCCCCCCCCCHHHHHHHHHCCCCC
AA: MQKYEKLEKIGEGTYGTVFKAKNRETHEIVALKRVRLDDDDEGVPSSALREICLLKELKH
10 20 30 40 50 60
Conf: 991246100223980899977235405677541899999788999999998333211137
Pred: CCEEEEEEEEECCCEEEEEEEECCCHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHCCC
AA: KNIVRLHDVLHSDKKLTLVFEFCDQDLKKYFDSCNGDLDPEIVKSFLFQLLKGLGFCHSR
70 80 90 100 110 120
Conf: 422678998420005799700132210111076443331003784414850003577767
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCEEEEEEECCCCCCCCCCCCC
AA: NVLHRDLKPQNLLINRNGELKLADFGLARAFGIPVRCYSAEVVTLWYRPPDVLFGAKLYS
130 140 150 160 170 180
Conf: 602556566799995637999889998256899989851999999899886799999999
Pred: CCHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC
AA: TSIDMWSAGCIFAELANAGRPLFPGNDVDDQLKRIFRLLGTPTEEQWPSMTKLPDYKPYP
190 200 210 220 230 240
Conf: 9988882323167899307999987554599988898888389997899999
Pred: CCCCCCCHHHHCCCCCHHHHHHHHHHHCCCCCCCCCHHHHHCCCCCCCCCCC
AA: MYPATTSLVNVVPKLNATGRDLLQNLLKCNPVQRISAEEALQHPYFSDFCPP
250 260 270 280 290
Show Conservation Score by AL2CO
1 M 0.573
2 Q -0.535
3 K -0.944
4 Y 1.148
5 E -0.996
6 K -0.281
7 L -0.573
8 E -0.712
9 K -0.666
10 I 0.288
11 G 0.462
12 E 0.287
13 G 2.208
14 T 0.618
15 Y 1.314
16 G 1.173
17 T -0.379
18 V 1.847
19 F 0.642
20 K 0.208
21 A 0.579
22 K -0.679
23 N -0.357
24 R -0.809
25 E -1.182
26 T -0.186
27 H -0.698
28 E -0.911
29 I -0.720
30 V -0.068
31 A 2.208
32 L 0.652
33 K 2.208
34 R -0.408
35 V 0.067
36 R -0.322
37 L 0.123
38 D -0.670
39 D -1.161
40 D -0.656
41 D -0.933
42 E -0.186
43 G 2.208
44 V -0.172
45 P 0.519
46 S -0.716
47 S 0.805
48 A 0.768
49 L 0.391
50 R 2.208
51 E 2.208
52 I 0.820
53 C 0.037
54 L 0.027
55 L 1.238
56 K -0.132
57 E -0.631
58 L 0.008
59 K -0.792
60 H 0.204
61 K -0.993
62 N -0.102
63 I 0.788
64 V 1.015
65 R -1.072
66 L 0.727
67 H -1.020
68 D 0.047
69 V 0.001
70 L -0.718
71 H -0.918
72 S -1.137
73 D -0.736
74 K -0.999
75 K -0.822
76 L 0.367
77 T -0.461
78 L 0.988
79 V 1.081
80 F 0.643
81 E 1.558
82 F 0.435
83 C -0.166
84 D -0.653
85 Q -1.192
86 D 1.848
87 L 1.038
88 K -0.068
89 K -0.882
90 Y -0.466
91 F -0.216
92 D -0.396
93 S -1.076
94 C -1.252
95 N -0.974
96 G -1.311
97 D -1.244
98 L -0.681
99 D -1.020
100 P -1.153
101 E -1.245
102 I -0.753
103 V -0.169
104 K 0.275
105 S -1.125
106 F -0.057
107 L -0.594
108 F -0.978
109 Q 0.522
110 L 0.420
111 L -0.276
112 K -0.962
113 G 1.197
114 L 0.302
115 G -1.115
116 F -0.033
117 C 0.427
118 H 2.208
119 S -1.168
120 R -0.223
121 N -0.572
122 V 0.761
123 L 0.489
124 H 2.208
125 R 1.844
126 D 2.208
127 L 1.031
128 K 1.848
129 P 0.503
130 Q 0.091
131 N 2.208
132 L 1.178
133 L 2.208
134 I 0.143
135 N -0.050
136 R -1.052
137 N -1.033
138 G -0.740
139 E -1.174
140 L 0.528
141 K 1.464
142 L 0.465
143 A 0.976
144 D 2.208
145 F 1.538
146 G 1.847
147 L 1.748
148 A 1.020
149 R 1.637
150 A -0.955
151 F -0.201
152 G -0.377
153 I -0.752
154 P 0.389
155 V -0.984
156 R -0.311
157 C -0.933
158 Y 0.327
159 S 1.173
160 A -0.336
161 E -0.335
162 V 1.206
163 V 0.600
164 T 1.620
165 L 0.715
166 W 1.495
167 Y 1.848
168 R 1.534
169 P 0.499
170 P 1.848
171 D 0.784
172 V 0.874
173 L 2.208
174 F 0.424
175 G 1.282
176 A -0.364
177 K -0.776
178 L -1.222
179 Y 2.208
180 S -0.133
181 T -0.704
182 S -0.265
183 I 0.442
184 D 2.208
185 M 0.008
186 W 2.208
187 S 1.049
188 A 0.015
189 G 1.510
190 C 0.666
191 I 1.372
192 F 0.186
193 A -0.431
194 E 2.208
195 L 0.354
196 A -0.654
197 N -0.889
198 A -0.587
199 G -0.027
200 R -0.944
201 P 0.163
202 L -0.113
203 F 0.727
204 P -0.814
205 G 1.596
206 N -0.849
207 D -0.217
208 V -0.002
209 D -1.072
210 D 0.115
211 Q 0.769
212 L 0.414
213 K -1.188
214 R -0.557
215 I 0.702
216 F -0.049
217 R -0.679
218 L -0.944
219 L -0.338
220 G 0.186
221 T 0.251
222 P 1.387
223 T -0.684
224 E -0.712
225 E -1.082
226 Q -0.908
227 W 0.225
228 P -0.469
229 S -0.499
230 M -0.538
231 T -1.064
232 K -1.044
233 L -0.540
234 P -0.376
235 D -0.692
236 Y -0.319
237 K -0.955
238 P -1.111
239 Y -1.137
240 P -0.993
241 M -1.205
242 Y -1.139
243 P -1.207
244 A -1.237
245 T -1.183
246 T -1.243
247 S -1.221
248 L -0.982
249 V -1.205
250 N -1.209
251 V -1.091
252 V -0.952
253 P -0.977
254 K -1.201
255 L -0.487
256 N -0.556
257 A -1.039
258 T -1.252
259 G -0.026
260 R -0.852
261 D -0.655
262 L 0.618
263 L 0.250
264 Q -1.087
265 N -0.915
266 L 0.025
267 L 0.347
268 K -1.029
269 C -0.330
270 N -0.932
271 P 0.187
272 V -1.125
273 Q -1.155
274 R 1.861
275 I -0.314
276 S -0.175
277 A 0.263
278 E -0.884
279 E -0.880
280 A -0.032
281 L -0.151
282 Q -1.176
283 H -0.356
284 P -1.027
285 Y 0.925
286 F 1.302
287 S -1.053
288 D -0.449
289 F -1.019
290 C -1.000 *
291 P -1.000 *
292 P -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 194.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MQKYEKLEKI GEGTYGTVFK AKNRETHEIV ALKRVRLDDD DEGVPSSALR EICLLKELKH
70 80 90 100 110 120
KNIVRLHDVL HSDKKLTLVF EFCDQDLKKY FDSCNGDLDP EIVKSFLFQL LKGLGFCHSR
130 140 150 160 170 180
NVLHRDLKPQ NLLINRNGEL KLADFGLARA FGIPVRCYSA EVVTLWYRPP DVLFGAKLYS
190 200 210 220 230 240
TSIDMWSAGC IFAELANAGR PLFPGNDVDD QLKRIFRLLG TPTEEQWPSM TKLPDYKPYP
250 260 270 280 290
MYPATTSLVN VVPKLNATGR DLLQNLLKCN PVQRISAEEA LQHPYFSDFC PP
3D Structures in PDB
1H4L (X-Ray,2.65 Å resolution)
1UNG (X-Ray,2.30 Å resolution)
Comments
Cdk5 is critical to the neuronal cell cycle and its nuclear localization is dependent upon interaction with p27. Cdk5 does not contain NLS of its own, but has two NES that direct it to the cytoplasm. Cdk5 nuclear export is required in postmitotic neurons for cell cycle re-entry, but once
in the cytoplasm, Cdk5 may protect against rapid neuronal death.
Doubts about NES: Neither proposed NES bound CRM1 in GST-pulldown assay (Chook Lab, unpublished results).
References
[1]. "Cdk5 nuclear localization is p27-dependent in nerve cells: implications for cell cycle suppression and caspase-3 activation."
Zhang J, Li H, Herrup K. (2010)
J Biol Chem,
285:14052-61
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
