Summary for Cdc7 (NES ID: 197)
Full Name
Cell division cycle 7-related protein kinase
UniProt Alternative Names
CDC7-related kinase, HsCdc7, huCdc7
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
Secondary Structure of Export Signal
α-helix
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|12643528|sp|O00311.1|CDC7_HUMAN RecName: Full=Cell division cycle 7-related protein kinase; Short=CDC7-related kinase; Short=HsCdc7; Short=huCdc7
MEASLGIQMDEPMAFSPQRDRFQAEGSLKKNEQNFKLAGVKKDIEKLYEAVPQLSNVFKIEDKIGEGTFS
SVYLATAQLQVGPEEKIALKHLIPTSHPIRIAAELQCLTVAGGQDNVMGVKYCFRKNDHVVIAMPYLEHE
SFLDILNSLSFQEVREYMLNLFKALKRIHQFGIVHRDVKPSNFLYNRRLKKYALVDFGLAQGTHDTKIEL
LKFVQSEAQQERCSQNKSHIITGNKIPLSGPVPKELDQQSTTKASVKRPYTNAQIQIKQGKDGKEGSVGL
SVQRSVFGERNFNIHSSISHESPAVKLMKQSKTVDVLSRKLATKKKAISTKVMNSAVMRKTASSCPASLT
CDCYATDKVCSICLSRRQQVAPRAGTPGFRAPEVLTKCPNQTTAIDMWSAGVIFLSLLSGRYPFYKASDD
LTALAQIMTIRGSRETIQAAKTFGKSILCSKEVPAQDLRKLCERLRGMDSSTPKLTSDIQGHASHQPAIS
EKTDHKASCLVQTPPGQYSGNSFKKGDSNSCEHCFDEYNTNLEGWNEVPDEAYDLLDKLLDLNPASRITA
EEALLHPFFKDMSL
Show Domain Info by CDD
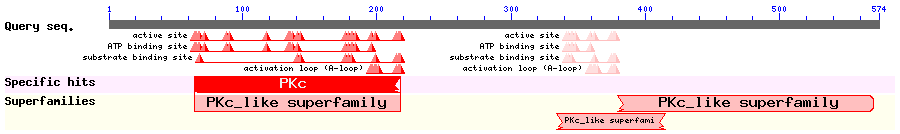
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 944358887898878967410000141112312465451499999998723899998589
Pred: CCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHCCCCCCCEEE
AA: MEASLGIQMDEPMAFSPQRDRFQAEGSLKKNEQNFKLAGVKKDIEKLYEAVPQLSNVFKI
10 20 30 40 50 60
Conf: 813303322179996750478996179997617898668899999998980899963473
Pred: EEEECCCCCEEEEEEEEECCCCCCCEEEEEEECCCCCHHHHHHHHHHHHHHCCCCCEEEE
AA: EDKIGEGTFSSVYLATAQLQVGPEEKIALKHLIPTSHPIRIAAELQCLTVAGGQDNVMGV
70 80 90 100 110 120
Conf: 577541995799940688996999971699899999999999999978628993345899
Pred: EEEEEECCEEEEEEECCCCCCHHHHHCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCC
AA: KYCFRKNDHVVIAMPYLEHESFLDILNSLSFQEVREYMLNLFKALKRIHQFGIVHRDVKP
130 140 150 160 170 180
Conf: 961021467904554200234567730122221123456632038875433589888999
Pred: CCCEEECCCCEEEEEEECCCCCCCCCCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCC
AA: SNFLYNRRLKKYALVDFGLAQGTHDTKIELLKFVQSEAQQERCSQNKSHIITGNKIPLSG
190 200 210 220 230 240
Conf: 999877777764357899986100100267999999877666644457665344578788
Pred: CCCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: PVPKELDQQSTTKASVKRPYTNAQIQIKQGKDGKEGSVGLSVQRSVFGERNFNIHSSISH
250 260 270 280 290 300
Conf: 880333104666410123220023345554565610003577899997776788887323
Pred: CCCHHHHCCCCCCCHHHHHHHCCCCCCCCCCCCCCHHCCCCCCCCCCCCCCCCCCCCCCC
AA: ESPAVKLMKQSKTVDVLSRKLATKKKAISTKVMNSAVMRKTASSCPASLTCDCYATDKVC
310 320 330 340 350 360
Conf: 123352346688777999888600017999973145567899999986378999878990
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHCCCCCCCCCCC
AA: SICLSRRQQVAPRAGTPGFRAPEVLTKCPNQTTAIDMWSAGVIFLSLLSGRYPFYKASDD
370 380 390 400 410 420
Conf: 899999999717798999999809710006887863189999997047789998754535
Pred: HHHHHHHHHHHCCHHHHHHHHHCCCCEECCCCCCCCHHHHHHHHHHCCCCCCCCCCCCCC
AA: LTALAQIMTIRGSRETIQAAKTFGKSILCSKEVPAQDLRKLCERLRGMDSSTPKLTSDIQ
430 440 450 460 470 480
Conf: 889989874322332223456899999889987679999876655656677678878832
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCH
AA: GHASHQPAISEKTDHKASCLVQTPPGQYSGNSFKKGDSNSCEHCFDEYNTNLEGWNEVPD
490 500 510 520 530 540
Conf: 5999998523689999999988844989678999
Pred: HHHHHHHHHCCCCCCCCCCHHHHHCCCCCCCCCC
AA: EAYDLLDKLLDLNPASRITAEEALLHPFFKDMSL
550 560 570
Show Conservation Score by AL2CO
1 M -1.000 *
2 E -1.000 *
3 A -1.000 *
4 S -1.000 *
5 L -1.000 *
6 G -1.000 *
7 I -1.000 *
8 Q -1.000 *
9 M -1.000 *
10 D -1.000 *
11 E -1.000 *
12 P -1.000 *
13 M -1.000 *
14 A -1.000 *
15 F -1.000 *
16 S -1.000 *
17 P -1.000 *
18 Q -1.000 *
19 R -1.000 *
20 D -1.000 *
21 R -1.000 *
22 F -1.000 *
23 Q -1.000 *
24 A -1.000 *
25 E -1.000 *
26 G -1.000 *
27 S -1.000 *
28 L -1.000 *
29 K -1.000 *
30 K -1.000 *
31 N -1.000 *
32 E -1.000 *
33 Q -1.000 *
34 N -1.000 *
35 F -1.000 *
36 K -1.000 *
37 L -1.000 *
38 A -1.000 *
39 G -1.000 *
40 V -1.000 *
41 K -1.000 *
42 K -1.000 *
43 D -1.000 *
44 I -1.000 *
45 E -1.000 *
46 K -1.000 *
47 L -1.000 *
48 Y -1.000 *
49 E -0.213
50 A -0.734
51 V -0.108
52 P 0.049
53 Q -0.198
54 L 0.470
55 S -0.415
56 N 0.138
57 V -0.409
58 F 2.895
59 K -0.146
60 I 0.681
61 E 0.162
62 D 0.323
63 K -0.071
64 I 2.186
65 G 2.047
66 E 0.056
67 G 4.714
68 T 1.449
69 F 2.526
70 S 2.510
71 S -0.297
72 V 2.641
73 Y 0.649
74 L 0.703
75 A 1.606
76 T 0.252
77 A -0.115
78 Q -0.070
79 L -0.543
80 Q -0.228
81 V -0.436
82 G -0.674
83 P -0.501
84 E -0.333
85 E -0.040
86 K -0.319
87 I 0.990
88 A 1.734
89 L 1.676
90 K 3.238
91 H -0.351
92 L 0.697
93 I -0.361
94 P 0.174
95 T -0.079
96 S 0.139
97 H -0.394
98 P -0.150
99 I -0.755
100 R 0.395
101 I 0.385
102 A -0.568
103 A -0.135
104 E 3.156
105 L 0.443
106 Q -0.294
107 C 0.435
108 L 1.614
109 T -0.406
110 V -0.547
111 A -0.103
112 G -0.090
113 G -0.160
114 Q -0.154
115 D 0.079
116 N 0.561
117 V 1.999
118 M 1.105
119 G 0.030
120 V 1.006
121 K -0.070
122 Y -0.110
123 C 0.614
124 F 0.978
125 R 0.255
126 K -0.425
127 N -0.292
128 D -0.023
129 H -0.216
130 V 0.364
131 V 1.149
132 I 1.210
133 A 2.048
134 M 1.496
135 P 2.028
136 Y 1.434
137 L 0.449
138 E -0.371
139 H 1.063
140 E 0.176
141 S 0.034
142 F 0.213
143 L -0.613
144 D -0.248
145 I 0.157
146 L 0.278
147 N -0.358
148 S -0.542
149 L 0.875
150 S 0.273
151 F -0.011
152 Q -0.608
153 E -0.388
154 V 1.047
155 R 0.324
156 E -0.724
157 Y 1.431
158 M 0.703
159 L -0.391
160 N 0.427
161 L 1.021
162 F 0.839
163 K -0.443
164 A 1.134
165 L 2.504
166 K 0.063
167 R -0.018
168 I 1.192
169 H 3.814
170 Q -0.041
171 F -0.196
172 G 0.332
173 I 1.767
174 V 0.968
175 H 3.707
176 R 2.364
177 D 4.714
178 V 2.107
179 K 3.210
180 P 2.110
181 S 0.824
182 N 3.911
183 F 1.944
184 L 2.042
185 Y 0.809
186 N 0.631
187 R -0.548
188 R -0.320
189 L -0.507
190 K 0.297
191 K -0.170
192 Y 0.311
193 A -0.054
194 L 2.225
195 V 1.007
196 D 2.917
197 F 1.348
198 G 2.019
199 L 1.060
200 A 1.970
201 Q 0.068
202 G -0.697
203 T -0.423
204 H -0.568
205 D -0.554
206 T -0.588
207 K -0.584
208 I -0.736
209 E -0.491
210 L -0.713
211 L -0.624
212 K -0.565
213 F -0.666
214 V -0.757
215 Q -0.585
216 S -0.569
217 E -0.657
218 A -0.661
219 Q -0.719
220 Q -0.666
221 E -0.568
222 R -0.703
223 C -0.769
224 S -0.567
225 Q -0.690
226 N -0.776
227 K -0.666
228 S -0.713
229 H -0.732
230 I -0.833
231 I -0.825
232 T -0.586
233 G -0.751
234 N -0.589
235 K -0.776
236 I -0.762
237 P -0.667
238 L -0.754
239 S -0.638
240 G -0.655
241 P -0.698
242 V -0.626
243 P -0.660
244 K -0.735
245 E -0.675
246 L -0.668
247 D -0.703
248 Q -0.749
249 Q -0.666
250 S -0.662
251 T -0.765
252 T -0.830
253 K -0.739
254 A -0.761
255 S -0.662
256 V -0.762
257 K -0.848
258 R -0.796
259 P -0.648
260 Y -0.861
261 T -0.736
262 N -0.645
263 A -0.790
264 Q -0.661
265 I -0.675
266 Q -0.738
267 I -0.834
268 K -0.649
269 Q -0.697
270 G -0.746
271 K -0.759
272 D -0.654
273 G -0.802
274 K -0.623
275 E -0.720
276 G -0.704
277 S -0.572
278 V -0.681
279 G -0.726
280 L -0.740
281 S -0.702
282 V -0.754
283 Q -0.687
284 R -0.597
285 S -0.730
286 V -0.620
287 F -0.801
288 G -0.737
289 E -0.693
290 R -0.593
291 N -0.556
292 F -0.762
293 N -0.725
294 I -0.760
295 H -0.808
296 S -0.729
297 S -0.537
298 I -0.674
299 S -0.662
300 H -0.642
301 E -0.591
302 S -0.710
303 P -0.812
304 A -0.801
305 V -0.759
306 K -0.759
307 L -0.869
308 M -0.807
309 K -0.570
310 Q -0.494
311 S -0.664
312 K -0.820
313 T -0.636
314 V -0.756
315 D -0.727
316 V -0.721
317 L -0.795
318 S -0.842
319 R -0.601
320 K -0.756
321 L -0.680
322 A -0.687
323 T -0.580
324 K -0.649
325 K -0.425
326 K -0.493
327 A -0.698
328 I -0.689
329 S -0.637
330 T -0.533
331 K -0.634
332 V -0.699
333 M -0.691
334 N -0.695
335 S -0.640
336 A -0.593
337 V -0.679
338 M -0.664
339 R -0.553
340 K -0.482
341 T -0.662
342 A -0.710
343 S -0.661
344 S -0.659
345 C -0.737
346 P -0.507
347 A -0.763
348 S -0.685
349 L -0.688
350 T -0.571
351 C -0.733
352 D -0.619
353 C -0.751
354 Y -0.776
355 A -0.631
356 T -0.651
357 D -0.487
358 K -0.707
359 V -0.597
360 C -0.579
361 S -0.664
362 I -0.794
363 C -0.766
364 L -0.618
365 S -0.525
366 R -0.399
367 R -0.629
368 Q -0.527
369 Q -0.685
370 V -0.706
371 A -0.202
372 P -0.378
373 R -0.531
374 A -0.439
375 G -0.285
376 T -0.105
377 P -0.107
378 G -0.055
379 F -0.493
380 R -0.526
381 A 0.046
382 P -0.186
383 E 0.408
384 V 0.643
385 L 0.496
386 T -0.534
387 K 0.046
388 C -0.542
389 P -0.470
390 N -0.672
391 Q -0.472
392 T 0.491
393 T -0.444
394 A -0.157
395 I 0.469
396 D 1.148
397 M 0.730
398 W 1.115
399 S 1.355
400 A 0.310
401 G 2.366
402 V 0.633
403 I 0.781
404 F 0.423
405 L 0.170
406 S 0.088
407 L 0.029
408 L 0.946
409 S -0.219
410 G 0.751
411 R -0.257
412 Y 0.126
413 P 2.015
414 F 0.784
415 Y 0.154
416 K -0.235
417 A 0.038
418 S -0.318
419 D 0.604
420 D 0.556
421 L -1.000 *
422 T -0.335
423 A 0.676
424 L 0.695
425 A -0.250
426 Q 0.024
427 I 1.381
428 M -0.306
429 T 0.059
430 I 0.180
431 R 0.351
432 G -1.000 *
433 S -1.000 *
434 R -1.000 *
435 E -1.000 *
436 T -1.000 *
437 I -1.000 *
438 Q -1.000 *
439 A -1.000 *
440 A -1.000 *
441 K -1.000 *
442 T -1.000 *
443 F -1.000 *
444 G -1.000 *
445 K -1.000 *
446 S -1.000 *
447 I -1.000 *
448 L -1.000 *
449 C -1.000 *
450 S -1.000 *
451 K -1.000 *
452 E -1.000 *
453 V -1.000 *
454 P -1.000 *
455 A -1.000 *
456 Q -1.000 *
457 D -1.000 *
458 L -1.000 *
459 R -1.000 *
460 K -1.000 *
461 L -1.000 *
462 C -1.000 *
463 E -1.000 *
464 R -1.000 *
465 L -1.000 *
466 R -1.000 *
467 G -1.000 *
468 M -1.000 *
469 D -1.000 *
470 S -1.000 *
471 S -1.000 *
472 T -1.000 *
473 P -1.000 *
474 K -1.000 *
475 L -1.000 *
476 T -1.000 *
477 S -1.000 *
478 D -1.000 *
479 I -1.000 *
480 Q -1.000 *
481 G -1.000 *
482 H -1.000 *
483 A -1.000 *
484 S -1.000 *
485 H -1.000 *
486 Q -1.000 *
487 P -1.000 *
488 A -1.000 *
489 I -1.000 *
490 S -1.000 *
491 E -1.000 *
492 K -1.000 *
493 T -1.000 *
494 D -1.000 *
495 H -1.000 *
496 K -1.000 *
497 A -1.000 *
498 S -1.000 *
499 C -1.000 *
500 L -1.000 *
501 V -1.000 *
502 Q -1.000 *
503 T -1.000 *
504 P -1.000 *
505 P -1.000 *
506 G -1.000 *
507 Q -1.000 *
508 Y -1.000 *
509 S -1.000 *
510 G -1.000 *
511 N -1.000 *
512 S -1.000 *
513 F -1.000 *
514 K -1.000 *
515 K -1.000 *
516 G -1.000 *
517 D -1.000 *
518 S -1.000 *
519 N -1.000 *
520 S -1.000 *
521 C -1.000 *
522 E -1.000 *
523 H -1.000 *
524 C -1.000 *
525 F -1.000 *
526 D -1.000 *
527 E -1.000 *
528 Y -1.000 *
529 N -1.000 *
530 T -1.000 *
531 N -1.000 *
532 L -1.000 *
533 E -1.000 *
534 G -1.000 *
535 W -1.000 *
536 N -1.000 *
537 E -1.000 *
538 V -1.000 *
539 P -1.000 *
540 D -1.000 *
541 E -1.000 *
542 A -1.000 *
543 Y -1.000 *
544 D -1.000 *
545 L -1.000 *
546 L -1.000 *
547 D -1.000 *
548 K -1.000 *
549 L -1.000 *
550 L -1.000 *
551 D -1.000 *
552 L -1.000 *
553 N -1.000 *
554 P -1.000 *
555 A -1.000 *
556 S -1.000 *
557 R -1.000 *
558 I -1.000 *
559 T -1.000 *
560 A -1.000 *
561 E -1.000 *
562 E -1.000 *
563 A -1.000 *
564 L -1.000 *
565 L -1.000 *
566 H -1.000 *
567 P -1.000 *
568 F -1.000 *
569 F -1.000 *
570 K -1.000 *
571 D -1.000 *
572 M -1.000 *
573 S -1.000 *
574 L -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 197.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MEASLGIQMD EPMAFSPQRD RFQAEGSLKK NEQNFKLAGV KKDIEKLYEA VPQLSNVFKI
70 80 90 100 110 120
EDKIGEGTFS SVYLATAQLQ VGPEEKIALK HLIPTSHPIR IAAELQCLTV AGGQDNVMGV
130 140 150 160 170 180
KYCFRKNDHV VIAMPYLEHE SFLDILNSLS FQEVREYMLN LFKALKRIHQ FGIVHRDVKP
190 200 210 220 230 240
SNFLYNRRLK KYALVDFGLA QGTHDTKIEL LKFVQSEAQQ ERCSQNKSHI ITGNKIPLSG
250 260 270 280 290 300
PVPKELDQQS TTKASVKRPY TNAQIQIKQG KDGKEGSVGL SVQRSVFGER NFNIHSSISH
310 320 330 340 350 360
ESPAVKLMKQ SKTVDVLSRK LATKKKAIST KVMNSAVMRK TASSCPASLT CDCYATDKVC
370 380 390 400 410 420
SICLSRRQQV APRAGTPGFR APEVLTKCPN QTTAIDMWSA GVIFLSLLSG RYPFYKASDD
430 440 450 460 470 480
LTALAQIMTI RGSRETIQAA KTFGKSILCS KEVPAQDLRK LCERLRGMDS STPKLTSDIQ
490 500 510 520 530 540
GHASHQPAIS EKTDHKASCL VQTPPGQYSG NSFKKGDSNS CEHCFDEYNT NLEGWNEVPD
550 560 570
EAYDLLDKLL DLNPASRITA EEALLHPFFK DMSL
3D Structures in PDB
5UWR (X-ray,2.24 Å resolution)
5UWQ (X-ray,2.28 Å resolution)
Comments
Cdc7 phosphorylates MCM, which is involved in the initiation of DNA replication. Cdc7 is reported to have an NLS, an NRS (nuclear retention sequence), as well as two NES. The NRS interacts with chromatin and is located within the purported NLS between amino acids 306-326. The two NES can mediate nuclear export at low efficiency, but both NES1 and 2 are required for an effective Cdc7 nucleocytoplasmic export.
References
[1]. "Identification and characterization of human cdc7 nuclear retention and export sequences in the context of chromatin binding."
Kim BJ, Kim SY, Lee H. (2007)
J Biol Chem,
282:30029-38
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
