NAD-dependent deacetylase sirtuin-2
UniProt
Show FASTA Format
>gi|38258608|sp|Q8IXJ6.2|SIRT2_HUMAN RecName: Full=NAD-dependent deacetylase sirtuin-2; AltName: Full=SIR2-like protein 2
MAEPDPSHPLETQAGKVQEAQDSDSDSEGGAAGGEADMDFLRNLFSQTLSLGSQKERLLDELTLEGVARY
MQSERCRRVICLVGAGISTSAGIPDFRSPSTGLYDNLEKYHLPYPEAIFEISYFKKHPEPFFALAKELYP
GQFKPTICHYFMRLLKDKGLLLRCYTQNIDTLERIAGLEQEDLVEAHGTFYTSHCVSASCRHEYPLSWMK
EKIFSEVTPKCEDCQSLVKPDIVFFGESLPARFFSCMQSDFLKVDLLLVMGTSLQVQPFASLISKAPLST
PRLLINKEKAGQSDPFLGMIMGLGGGMDFDSKKAYRDVAWLGECDQGCLALAELLGWKKELEDLVRREHA
SIDAQSGAGVPNPSTSASPKKSPPPAKDEARTTEREKPQ
Show Domain Info by CDD
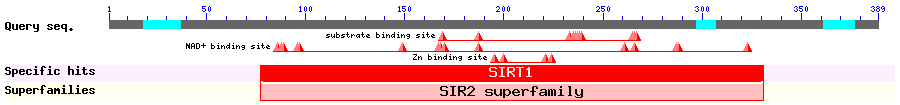
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 999999999565545222234899987789899974079999999976128987655567
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCC
AA: MAEPDPSHPLETQAGKVQEAQDSDSDSEGGAAGGEADMDFLRNLFSQTLSLGSQKERLLD
10 20 30 40 50 60
Conf: 689899999987089853899706852357989998789985122367899999653110
Pred: CCCHHHHHHHHHHCCCCEEEEEECCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: ELTLEGVARYMQSERCRRVICLVGAGISTSAGIPDFRSPSTGLYDNLEKYHLPYPEAIFE
70 80 90 100 110 120
Conf: 344234994489987664699989992358899764128500001367623577409998
Pred: CCCCCCCCHHHHHHHHHHCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHCCCC
AA: ISYFKKHPEPFFALAKELYPGQFKPTICHYFMRLLKDKGLLLRCYTQNIDTLERIAGLEQ
130 140 150 160 170 180
Conf: 782552264224655622249988938899985239998689999942586002399980
Pred: CCEEEECCCCCCCCCCCCCCCCCCCHHHHHHHHHCCCCCCCCCCCCCCCCCEEEECCCCH
AA: EDLVEAHGTFYTSHCVSASCRHEYPLSWMKEKIFSEVTPKCEDCQSLVKPDIVFFGESLP
190 200 210 220 230 240
Conf: 667975640888886899956564224464333569999994886155689999602332
Pred: HHHHHHHHCCCCCCCEEEEECCCCCCCCCCCCCCCCCCCCCEEEEECCCCCCCCCCHHHH
AA: ARFFSCMQSDFLKVDLLLVMGTSLQVQPFASLISKAPLSTPRLLINKEKAGQSDPFLGMI
250 260 270 280 290 300
Conf: 169998767898864540312775899999999909936888999853200002358999
Pred: CCCCCCCCCCCCCCCCCEEECCCHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCCCCCCC
AA: MGLGGGMDFDSKKAYRDVAWLGECDQGCLALAELLGWKKELEDLVRREHASIDAQSGAGV
310 320 330 340 350 360
Conf: 99999989999999964223564446999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: PNPSTSASPKKSPPPAKDEARTTEREKPQ
370 380
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 E -1.000 *
4 P -1.000 *
5 D -1.000 *
6 P -1.000 *
7 S -1.000 *
8 H -1.000 *
9 P -1.000 *
10 L -1.000 *
11 E -1.000 *
12 T -1.000 *
13 Q -1.000 *
14 A -1.000 *
15 G -1.000 *
16 K -1.000 *
17 V -1.000 *
18 Q -1.000 *
19 E -1.000 *
20 A -1.000 *
21 Q -1.000 *
22 D -1.000 *
23 S -1.000 *
24 D -1.000 *
25 S -1.000 *
26 D -1.000 *
27 S -1.000 *
28 E -1.000 *
29 G -1.000 *
30 G -1.000 *
31 A -1.000 *
32 A -1.000 *
33 G -1.000 *
34 G -1.000 *
35 E -1.000 *
36 A -1.000 *
37 D -1.000 *
38 M -1.000 *
39 D -1.000 *
40 F -1.000 *
41 L -1.000 *
42 R -1.000 *
43 N -1.000 *
44 L -1.000 *
45 F -1.000 *
46 S -1.000 *
47 Q -1.000 *
48 T -1.000 *
49 L -1.000 *
50 S -0.863
51 L -1.006
52 G -0.921
53 S -0.863
54 Q -0.994
55 K -0.697
56 E -0.769
57 R -0.665
58 L -0.706
59 L -0.829
60 D -0.682
61 E -0.949
62 L -0.980
63 T -0.216
64 L 0.265
65 E -0.578
66 G -0.521
67 V 0.439
68 A -0.276
69 R -0.545
70 Y -0.784
71 M 0.461
72 Q -0.531
73 S -0.640
74 E -0.751
75 R -0.672
76 C -0.841
77 R -0.484
78 R 0.159
79 V 1.027
80 I 0.014
81 C 0.025
82 L 0.545
83 V -0.068
84 G 1.747
85 A 1.784
86 G 1.776
87 I 0.578
88 S 2.530
89 T 1.009
90 S 0.302
91 A 0.894
92 G 1.578
93 I 1.686
94 P 2.158
95 D 2.502
96 F 2.502
97 R 2.903
98 S 1.761
99 P 0.311
100 S -0.257
101 T -0.395
102 G 2.539
103 L 0.663
104 Y 2.024
105 D -0.306
106 N -0.076
107 L 0.560
108 E -0.469
109 K -0.711
110 Y 0.007
111 H -0.099
112 L 0.959
113 P 0.098
114 Y -0.385
115 P 1.485
116 E -0.004
117 A 0.305
118 I 0.840
119 F 2.903
120 E 0.087
121 I 0.393
122 S -0.489
123 Y 0.264
124 F 1.714
125 K -0.954
126 K -0.915
127 H -0.191
128 P 1.782
129 E -0.865
130 P 0.359
131 F 2.259
132 F 0.829
133 A -0.827
134 L 0.454
135 A 0.047
136 K -0.563
137 E -0.552
138 L 0.548
139 Y -0.436
140 P 0.343
141 G -0.379
142 Q -0.803
143 F -0.500
144 K -0.996
145 P 0.682
146 T 0.901
147 I -0.782
148 C -0.315
149 H 2.571
150 Y -0.713
151 F 1.692
152 M 0.094
153 R -0.115
154 L -0.455
155 L 0.519
156 K -0.645
157 D -0.567
158 K -0.269
159 G -0.136
160 L -0.515
161 L 2.144
162 L -0.591
163 R -0.091
164 C -0.253
165 Y 0.774
166 T 1.548
167 Q 2.117
168 N 2.188
169 I 1.368
170 D 2.522
171 T -0.102
172 L 1.725
173 E 1.854
174 R -0.793
175 I -0.808
176 A 0.649
177 G 0.441
178 L 0.674
179 E -0.609
180 Q -0.677
181 E -0.353
182 D -0.761
183 L 0.482
184 V 0.747
185 E -0.474
186 A 0.492
187 H 2.552
188 G 2.551
189 T 0.195
190 F 0.291
191 Y -0.609
192 T -0.675
193 S 0.097
194 H -0.302
195 C 2.539
196 V -0.345
197 S -0.772
198 A -0.250
199 S -0.810
200 C -1.000 *
201 R -0.953
202 H -0.810
203 E -0.809
204 Y -1.006
205 P -0.885
206 L -1.121
207 S -0.789
208 W -1.094
209 M -0.865
210 K -0.916
211 E -0.783
212 K -1.017
213 I -0.675
214 F -0.918
215 S -0.712
216 E -0.872
217 V -1.043
218 T -0.422
219 P 0.089
220 K -0.836
221 C 1.551
222 E -0.887
223 D -0.868
224 C -0.264
225 Q -0.985
226 S -0.101
227 L -0.394
228 V 0.630
229 K 1.654
230 P 1.262
231 D -0.206
232 I 0.466
233 V 0.629
234 F 0.896
235 F 0.759
236 G 0.626
237 E 1.096
238 S -0.562
239 L 0.931
240 P 0.717
241 A -0.937
242 R -0.778
243 F 0.596
244 F -1.024
245 S -0.935
246 C -1.015
247 M -0.819
248 Q -1.075
249 S -0.916
250 D -0.560
251 F -0.768
252 L -0.935
253 K -0.967
254 V -0.389
255 D 0.341
256 L 0.211
257 L -0.078
258 L 0.353
259 V 0.600
260 M 0.016
261 G 2.202
262 T 1.552
263 S 1.093
264 L 1.467
265 Q -0.525
266 V 1.248
267 Q -0.618
268 P 2.903
269 F 0.552
270 A 0.529
271 S -0.685
272 L 0.653
273 I -0.102
274 S -0.981
275 K -1.056
276 A -0.118
277 P -0.747
278 L -1.051
279 S -0.711
280 T -0.300
281 P -0.375
282 R 0.468
283 L 0.284
284 L 0.652
285 I 0.551
286 N 1.857
287 K -0.166
288 E 0.096
289 K -0.908
290 A -0.351
291 G -0.364
292 Q -0.907
293 S -1.000 *
294 D -1.000 *
295 P -1.000 *
296 F -1.000 *
297 L -1.000 *
298 G -1.000 *
299 M -1.000 *
300 I -1.000 *
301 M -1.000 *
302 G -1.000 *
303 L -1.000 *
304 G -1.000 *
305 G -1.000 *
306 G -1.000 *
307 M -1.000 *
308 D -1.000 *
309 F -1.000 *
310 D -0.810
311 S -1.164
312 K -0.816
313 K -0.900
314 A -0.809
315 Y -1.048
316 R -0.662
317 D 1.220
318 V -0.116
319 A -0.935
320 W -0.689
321 L -0.702
322 G -0.107
323 E -0.527
324 C 0.068
325 D -0.157
326 Q -0.865
327 G -0.703
328 C 0.214
329 L -1.019
330 A -0.814
331 L 0.608
332 A -0.092
333 E -0.617
334 L -0.815
335 L 0.489
336 G 0.005
337 W -0.093
338 K -0.876
339 K -1.026
340 E -0.772
341 L -0.788
342 E -0.870
343 D -0.817
344 L -0.698
345 V -0.965
346 R -1.012
347 R -0.635
348 E -0.901
349 H -1.122
350 A -0.932
351 S -0.824
352 I -0.936
353 D -0.853
354 A -0.975
355 Q -0.890
356 S -0.992
357 G -0.870
358 A -1.000 *
359 G -1.000 *
360 V -1.000 *
361 P -1.000 *
362 N -1.000 *
363 P -1.000 *
364 S -1.000 *
365 T -1.000 *
366 S -1.000 *
367 A -1.000 *
368 S -1.000 *
369 P -1.000 *
370 K -1.000 *
371 K -1.000 *
372 S -1.000 *
373 P -1.000 *
374 P -1.000 *
375 P -1.000 *
376 A -1.000 *
377 K -1.000 *
378 D -1.000 *
379 E -1.000 *
380 A -1.000 *
381 R -1.000 *
382 T -1.000 *
383 T -1.000 *
384 E -1.000 *
385 R -1.000 *
386 E -1.000 *
387 K -1.000 *
388 P -1.000 *
389 Q -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 199.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MAEPDPSHPL ETQAGKVQEA QDSDSDSEGG AAGGEADMDF LRNLFSQTLS LGSQKERLLD
70 80 90 100 110 120
ELTLEGVARY MQSERCRRVI CLVGAGISTS AGIPDFRSPS TGLYDNLEKY HLPYPEAIFE
130 140 150 160 170 180
ISYFKKHPEP FFALAKELYP GQFKPTICHY FMRLLKDKGL LLRCYTQNID TLERIAGLEQ
190 200 210 220 230 240
EDLVEAHGTF YTSHCVSASC RHEYPLSWMK EKIFSEVTPK CEDCQSLVKP DIVFFGESLP
250 260 270 280 290 300
ARFFSCMQSD FLKVDLLLVM GTSLQVQPFA SLISKAPLST PRLLINKEKA GQSDPFLGMI
310 320 330 340 350 360
MGLGGGMDFD SKKAYRDVAW LGECDQGCLA LAELLGWKKE LEDLVRREHA SIDAQSGAGV
370 380
PNPSTSASPK KSPPPAKDEA RTTEREKPQ
1J8F (X-Ray,1.70 Å resolution)
5FYQ ( X-ray ,3.00 Å resolution)
A tubulin deacetylase in the cytoplasm, SIRT2 contains an NES between amino acids 41-51. During mitosis, SIRT2 was found to localize to the centrosome, and overexpression of SIRT2 effects cell division and leads to multinucleation. SIRT2 might be imported into the nucleus through undefined NLS or through another protein. Sendino et al. showed that NES in 37-55 has export activity when tested in Rev1.4-GFP and SRV
A/B reporters and responded to CRM1 co-expression. They also identified another putative NES in 244-267, but the activity is weak and CRM1-dependency is not clearly illustrated.
Ref.2Using a Simple Cellular Assay to Map NES Motifs in Cancer-Related Proteins, Gain Insight into CRM1-Mediated NES Export, and Search for NES-Harboring Micropeptides, Sendino et al., Int J Mol Sci, 2020
[1]. "Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis."
North BJ, Verdin E. (2007)
PLoS One,
2:e784
PubMed[2]. "Using a Simple Cellular Assay to Map NES Motifs in Cancer-Related Proteins, Gain Insight into CRM1-Mediated NES Export, and Search for NES-Harboring Micropeptides"
Sendino M, Omaetxebarria MJ, Prieto G, Rodriguez JA. (2020)
Int J Mol Sci,
21(17):E6341
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
