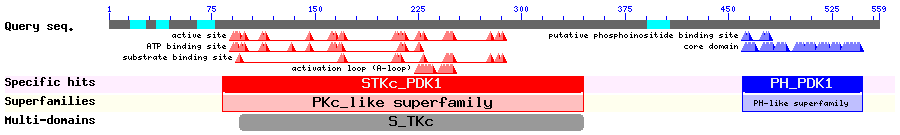
3-phosphoinositide-dependent protein kinase 1
UniProt
Show FASTA Format
>gi|18203660|sp|Q9Z2A0.2|PDPK1_MOUSE RecName: Full=3-phosphoinositide-dependent protein kinase 1; Short=mPDK1
MARTTSQLYDAVPIQSSVVLCSCPSPSMVRSQTEPGSSPGIPSGVSRQGSTMDGTTAEARPSTNPLQQHP
AQLPPQPRKKRPEDFKFGKILGEGSFSTVVLARELATSREYAIKILEKRHIIKENKVPYVTRERDVMSRL
DHPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIGSFDETCTRFYTAEIVSALEYLHGKGIIHRDLK
PENILLNEDMHIQITDFGTAKVLSPESKQARANSFVGTAQYVSPELLTEKSACKSSDLWALGCIIYQLVA
GLPPFRAGNEYLIFQKIIKLEYHFPEKFFPKARDLVEKLLVLDATKRLGCEEMEGYGPLKAHPFFETITW
ENLHQQTPPKLTAYLPAMSEDDEDCYGNYDNLLSQFGFMQVSSSSSSHSLSTVETSLPQRSGSNIEQYIH
DLDTNSFELDLQFSEDEKRLLLEKQAGGNPWHQFVENNLILKMGPVDKRKGLFARRRQLLLTEGPHLYYV
DPVNKVLKGEIPWSQELRPEAKNFKTFFVHTPNRTYYLMDPSGNAHKWCRKIQEVWRQQYQSNPDAAVQ
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 987555666776779983115899999767899999999999987889999999975679
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: MARTTSQLYDAVPIQSSVVLCSCPSPSMVRSQTEPGSSPGIPSGVSRQGSTMDGTTAEAR
10 20 30 40 50 60
Conf: 999976668999999898889787423114302543038999983438689998621001
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCEEEECCCCCEEEEEEECCCCHHHHHHHHHHHH
AA: PSTNPLQQHPAQLPPQPRKKRPEDFKFGKILGEGSFSTVVLARELATSREYAIKILEKRH
70 80 90 100 110 120
Conf: 242167205889999872029994541355430799417888504798225888852587
Pred: HHHHCCCHHHHHHHHHHHHCCCCCEEEEEEEEECCCCEEEEEEECCCCCHHHHHHHHCCC
AA: IIKENKVPYVTRERDVMSRLDHPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIGSF
130 140 150 160 170 180
Conf: 713467899999999998870996134699862012899968871045533479987578
Pred: CHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCEEEEECCCCCCCCCCCCCC
AA: DETCTRFYTAEIVSALEYLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQA
190 200 210 220 230 240
Conf: 876642346665831258899876511677889999995289996407867999999816
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHC
AA: RANSFVGTAQYVSPELLTEKSACKSSDLWALGCIIYQLVAGLPPFRAGNEYLIFQKIIKL
250 260 270 280 290 300
Conf: 656899999558999984112683632799877897455459986789957534789997
Pred: CCCCCCCCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: EYHFPEKFFPKARDLVEKLLVLDATKRLGCEEMEGYGPLKAHPFFETITWENLHQQTPPK
310 320 330 340 350 360
Conf: 656888999998755699998665567665567998998897678888999998655313
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: LTAYLPAMSEDDEDCYGNYDNLLSQFGFMQVSSSSSSHSLSTVETSLPQRSGSNIEQYIH
370 380 390 400 410 420
Conf: 589984234555686898643101259999875557971320154455556532120344
Pred: CCCCCCCCCCCCCCHHHHHHHHCCCCCCCCCCCCCCCCEEEECCCCCCCCCCCCCCCEEE
AA: DLDTNSFELDLQFSEDEKRLLLEKQAGGNPWHQFVENNLILKMGPVDKRKGLFARRRQLL
430 440 450 460 470 480
Conf: 306981899779985010001357544320118520899849904999879998999999
Pred: EECCCEEEEECCCCCEEEECCCCCCCCCCCCCCCCCEEEECCCEEEEEECCCCCHHHHHH
AA: LTEGPHLYYVDPVNKVLKGEIPWSQELRPEAKNFKTFFVHTPNRTYYLMDPSGNAHKWCR
490 500 510 520 530 540
Conf: 9999999984029986789
Pred: HHHHHHHHHHHCCCCCCCC
AA: KIQEVWRQQYQSNPDAAVQ
550
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 R -1.000 *
4 T -1.000 *
5 T -1.000 *
6 S -1.000 *
7 Q -1.000 *
8 L -1.000 *
9 Y -1.000 *
10 D -1.000 *
11 A -1.000 *
12 V -1.000 *
13 P -1.000 *
14 I -1.000 *
15 Q -1.000 *
16 S -1.000 *
17 S -1.000 *
18 V -1.000 *
19 V -1.000 *
20 L -1.000 *
21 C -1.000 *
22 S -1.000 *
23 C -1.000 *
24 P -1.000 *
25 S -1.000 *
26 P -1.000 *
27 S -1.000 *
28 M -1.000 *
29 V -1.000 *
30 R -1.000 *
31 S -1.000 *
32 Q -1.000 *
33 T -1.000 *
34 E -1.000 *
35 P -1.000 *
36 G -1.000 *
37 S -1.000 *
38 S -1.000 *
39 P -1.000 *
40 G -1.000 *
41 I -1.000 *
42 P -1.000 *
43 S -1.000 *
44 G -1.000 *
45 V -1.000 *
46 S -1.000 *
47 R -1.000 *
48 Q -1.000 *
49 G -1.000 *
50 S -1.000 *
51 T -1.000 *
52 M -1.122
53 D -0.976
54 G -1.038
55 T -1.032
56 T -1.067
57 A -1.139
58 E -1.005
59 A -1.150
60 R -1.006
61 P -1.040
62 S -1.012
63 T -1.134
64 N -1.013
65 P -1.008
66 L -1.057
67 Q -1.046
68 Q -1.080
69 H -1.109
70 P -1.029
71 A -1.040
72 Q -1.095
73 L -1.065
74 P -1.038
75 P -0.986
76 Q -0.945
77 P -0.928
78 R -0.889
79 K -0.550
80 K -0.821
81 R -0.645
82 P -0.388
83 E -0.799
84 D -0.073
85 F 1.515
86 K -0.894
87 F -0.191
88 G -0.672
89 K -0.134
90 I -0.086
91 L 1.027
92 G 2.660
93 E -0.039
94 G 2.265
95 S 0.412
96 F 1.544
97 S 1.364
98 T -0.097
99 V 2.245
100 V -0.412
101 L -0.183
102 A 0.375
103 R 0.170
104 E -0.611
105 L -0.149
106 A -0.979
107 T -0.189
108 S -0.599
109 R -0.927
110 E -0.683
111 Y 0.557
112 A 2.265
113 I 0.736
114 K 2.660
115 I 0.244
116 L 0.736
117 E -0.587
118 K 1.575
119 R -0.670
120 H -0.935
121 I 1.126
122 I -0.370
123 K -0.302
124 E -0.369
125 N 0.017
126 K 0.050
127 V -0.376
128 P -0.721
129 Y 0.129
130 V 0.835
131 T -0.514
132 R -0.558
133 E 2.660
134 R 0.739
135 D -0.623
136 V 0.480
137 M 1.225
138 S -0.758
139 R -0.957
140 L -0.463
141 D -0.675
142 H 0.079
143 P 0.488
144 F 0.535
145 F 0.739
146 V 0.579
147 K -0.685
148 L 1.246
149 Y -0.648
150 F -0.249
151 T 0.483
152 F 0.795
153 Q 0.792
154 D 0.805
155 D -0.874
156 E -0.836
157 K -0.248
158 L 1.227
159 Y 1.139
160 F 0.589
161 G 0.431
162 L 0.553
163 S 0.351
164 Y 0.270
165 A 0.148
166 K -0.781
167 N 0.420
168 G 2.264
169 E 0.703
170 L 1.293
171 L 0.901
172 K -0.920
173 Y -0.281
174 I 0.932
175 R -0.573
176 K -0.422
177 I -0.840
178 G -0.323
179 S -0.812
180 F 1.512
181 D -0.581
182 E -0.839
183 T -0.773
184 C -0.626
185 T 0.589
186 R -0.174
187 F 0.214
188 Y 1.175
189 T 0.001
190 A 0.715
191 E 1.242
192 I 0.800
193 V 0.035
194 S -0.559
195 A 0.637
196 L 0.740
197 E -0.464
198 Y -0.179
199 L 0.843
200 H 1.895
201 G -0.849
202 K -0.778
203 G -0.427
204 I 0.996
205 I 0.825
206 H 1.770
207 R 2.247
208 D 2.660
209 L 1.159
210 K 2.660
211 P 1.807
212 E 1.612
213 N 2.660
214 I 1.074
215 L 1.353
216 L 0.625
217 N 0.433
218 E -0.932
219 D -0.791
220 M 0.632
221 H 0.560
222 I 0.778
223 Q -0.274
224 I 1.025
225 T 0.853
226 D 2.660
227 F 2.087
228 G 2.660
229 T 0.351
230 A 0.770
231 K 0.764
232 V -0.746
233 L -0.675
234 S -0.819
235 P -0.874
236 E -0.767
237 S -1.038
238 K -1.154
239 Q -0.921
240 A -0.772
241 R -0.731
242 A -0.314
243 N -0.869
244 S 1.533
245 F 0.476
246 V 1.327
247 G 1.884
248 T 2.245
249 A 0.242
250 Q 0.574
251 Y 1.646
252 V 0.896
253 S 1.144
254 P 2.660
255 E 2.248
256 L 0.783
257 L 1.098
258 T -0.688
259 E -0.390
260 K -0.822
261 S -0.599
262 A -0.268
263 C -0.235
264 K -0.818
265 S -0.460
266 S 0.118
267 D 2.269
268 L 0.628
269 W 1.715
270 A 0.409
271 L 0.187
272 G 2.213
273 C 0.232
274 I 0.338
275 I 0.470
276 Y 1.278
277 Q 1.216
278 L 1.199
279 V -0.226
280 A -0.300
281 G 2.263
282 L -0.758
283 P 0.195
284 P 1.257
285 F 1.361
286 R -0.551
287 A -0.129
288 G -0.663
289 N 0.080
290 E -0.726
291 Y -0.894
292 L -0.722
293 I 0.297
294 F 0.443
295 Q -0.272
296 K -0.133
297 I 1.224
298 I -0.227
299 K -0.876
300 L -0.636
301 E -0.373
302 Y 0.407
303 H -0.928
304 F 0.151
305 P 0.095
306 E -0.836
307 K -0.958
308 F 0.007
309 F -0.766
310 P -0.873
311 K -0.914
312 A -0.022
313 R -0.485
314 D -0.352
315 L 0.847
316 V 0.748
317 E -0.679
318 K -0.435
319 L 1.186
320 L 0.916
321 V -0.837
322 L -0.609
323 D -0.024
324 A -0.467
325 T -0.955
326 K -0.650
327 R 2.247
328 L 0.191
329 G 0.917
330 C -0.288
331 E -0.812
332 E -0.722
333 M -1.000 *
334 E -0.720
335 G 0.425
336 Y -0.503
337 G -0.899
338 P -0.595
339 L 0.925
340 K -0.173
341 A -0.920
342 H 1.038
343 P -0.832
344 F 0.872
345 F 0.858
346 E -0.797
347 T -0.790
348 I 0.688
349 T -0.553
350 W 1.125
351 E -0.880
352 N -0.726
353 L 0.539
354 H -1.132
355 Q -0.873
356 Q -0.498
357 T -0.756
358 P -0.088
359 P -1.000 *
360 K -0.843
361 L -0.899
362 T -0.725
363 A -0.517
364 Y -0.654
365 L -0.903
366 P -0.208
367 A -0.942
368 M -0.897
369 S -0.968
370 E -0.955
371 D -0.845
372 D -1.113
373 E -0.771
374 D -0.846
375 C -1.118
376 Y -0.945
377 G -0.746
378 N -0.809
379 Y -0.657
380 D -0.510
381 N -0.876
382 L -1.088
383 L -0.878
384 S -0.727
385 Q -0.684
386 F -0.925
387 G -0.892
388 F -1.137
389 M -1.133
390 Q -0.994
391 V -0.968
392 S -0.952
393 S -1.054
394 S -1.123
395 S -0.991
396 S -1.047
397 S -1.081
398 H -1.101
399 S -1.000 *
400 L -1.000 *
401 S -1.020
402 T -1.000 *
403 V -1.000 *
404 E -1.000 *
405 T -1.000 *
406 S -1.000 *
407 L -1.000 *
408 P -1.000 *
409 Q -1.000 *
410 R -1.000 *
411 S -1.000 *
412 G -1.000 *
413 S -1.000 *
414 N -1.000 *
415 I -1.000 *
416 E -1.000 *
417 Q -1.000 *
418 Y -1.000 *
419 I -1.000 *
420 H -1.000 *
421 D -1.000 *
422 L -1.000 *
423 D -1.000 *
424 T -1.000 *
425 N -1.000 *
426 S -1.000 *
427 F -1.000 *
428 E -1.000 *
429 L -1.000 *
430 D -1.000 *
431 L -1.000 *
432 Q -1.000 *
433 F -1.000 *
434 S -1.000 *
435 E -1.000 *
436 D -1.000 *
437 E -1.000 *
438 K -1.000 *
439 R -1.000 *
440 L -1.000 *
441 L -1.000 *
442 L -1.000 *
443 E -1.000 *
444 K -1.000 *
445 Q -1.000 *
446 A -1.000 *
447 G -1.000 *
448 G -1.000 *
449 N -1.000 *
450 P -1.000 *
451 W -1.000 *
452 H -1.000 *
453 Q -1.000 *
454 F -1.000 *
455 V -1.000 *
456 E -1.000 *
457 N -1.000 *
458 N -1.000 *
459 L -1.000 *
460 I -1.000 *
461 L -1.000 *
462 K -1.000 *
463 M -1.000 *
464 G -1.000 *
465 P -1.000 *
466 V -1.000 *
467 D -1.000 *
468 K -1.000 *
469 R -1.000 *
470 K -1.000 *
471 G -1.000 *
472 L -1.000 *
473 F -1.000 *
474 A -1.000 *
475 R -1.000 *
476 R -1.000 *
477 R -1.000 *
478 Q -1.000 *
479 L -1.000 *
480 L -1.000 *
481 L -1.000 *
482 T -1.000 *
483 E -1.000 *
484 G -1.000 *
485 P -1.000 *
486 H -1.000 *
487 L -1.000 *
488 Y -1.000 *
489 Y -1.000 *
490 V -1.000 *
491 D -1.000 *
492 P -1.000 *
493 V -1.000 *
494 N -1.000 *
495 K -1.000 *
496 V -1.000 *
497 L -1.000 *
498 K -1.000 *
499 G -1.000 *
500 E -1.000 *
501 I -1.000 *
502 P -1.000 *
503 W -1.000 *
504 S -1.000 *
505 Q -1.000 *
506 E -1.000 *
507 L -1.000 *
508 R -1.000 *
509 P -1.000 *
510 E -1.000 *
511 A -1.000 *
512 K -1.000 *
513 N -1.000 *
514 F -1.000 *
515 K -1.000 *
516 T -1.000 *
517 F -1.000 *
518 F -1.000 *
519 V -1.000 *
520 H -1.000 *
521 T -1.000 *
522 P -1.000 *
523 N -1.000 *
524 R -1.000 *
525 T -1.000 *
526 Y -1.000 *
527 Y -1.000 *
528 L -1.000 *
529 M -1.000 *
530 D -1.000 *
531 P -1.000 *
532 S -1.000 *
533 G -1.000 *
534 N -1.000 *
535 A -1.000 *
536 H -1.000 *
537 K -1.000 *
538 W -1.000 *
539 C -1.000 *
540 R -1.000 *
541 K -1.000 *
542 I -1.000 *
543 Q -1.000 *
544 E -1.000 *
545 V -1.000 *
546 W -1.000 *
547 R -1.000 *
548 Q -1.000 *
549 Q -1.000 *
550 Y -1.000 *
551 Q -1.000 *
552 S -1.000 *
553 N -1.000 *
554 P -1.000 *
555 D -1.000 *
556 A -1.000 *
557 A -1.000 *
558 V -1.000 *
559 Q -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 200.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MARTTSQLYD AVPIQSSVVL CSCPXPSMVR SQTEPGSSPG IPSGVSRQGS TMDGTTAEAR
70 80 90 100 110 120
PSTNPLQQHP AQLPPQPRKK RPENFKFGKI LGEGSFSTVV LARELATSRE YAIKILEKRH
130 140 150 160 170 180
IIKENKVPYV TRERDVMSRL DHPFFVKLYF TFQDDEKLYF GLSYAKNGEL LKYIRKIGSF
190 200 210 220 230 240
DETCTRFYTA EIVSALEYLH GKGIIHRDLK PENILLNEDM HIQITDFGTA KVLSPESKQA
250 260 270 280 290 300
RANSFVGTAQ YVSPELLTEK SACKSSDLWA LGCIIYQLVA GLPPFRAGNE YLIFQKIIKL
310 320 330 340 350 360
EYHFPEKFFP KARDLVEKLL VLDATKRLGC EEMEGYGPLK AHPFFETITW ENLHQQTPPK
370 380 390 400 410 420
LTAYLPAMSE DDEDCYGNYD NLLSQFGFMQ VSSSSSSHSL STVETSLPQR SGSNIEQYIH
430 440 450 460 470 480
DLDTNSFELD LQFSEDEKRL LLEKQAGGNP WHQFVENNLI LKMGPVDKRK GLFARRRQLL
490 500 510 520 530 540
LTEGPHLYYV DPVNKVLKGE IPWSQELRPE AKNFKTFFVH TPNRTYYLMD PSGNAHKWCR
550
KIQEVWRQQY QSNPDAAVQ
PDK1 phosphorylates members of the AGC protein kinase family and is important in regulating cell survival. Lim et al. reported that PDK1 shuttles in LMB sensitive manner and deletion of aa 382-391 caused nuclear accumulation, as well as mutations of two hydrophobic residues in that region. PDK's nuclear translocation was regulated by PI3 pathway.
[1]. "Nuclear translocation of 3'-phosphoinositide-dependent protein kinase 1 (PDK-1): a potential regulatory mechanism for PDK-1 function"
Lim MA, Kikani CK, Wick MJ, Dong LQ. (2003)
Proc Natl Acad Sci,
100:14006-11
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
