NAD-dependent deacetylase HST2
UniProt
Show FASTA Format
>gi|1708326|sp|P53686.1|HST2_YEAST RecName: Full=NAD-dependent deacetylase HST2; AltName: Full=Homologous to SIR2 protein 2
MSVSTASTEMSVRKIAAHMKSNPNAKVIFMVGAGISTSCGIPDFRSPGTGLYHNLARLKLPYPEAVFDVD
FFQSDPLPFYTLAKELYPGNFRPSKFHYLLKLFQDKDVLKRVYTQNIDTLERQAGVKDDLIIEAHGSFAH
CHCIGCGKVYPPQVFKSKLAEHPIKDFVKCDVCGELVKPAIVFFGEDLPDSFSETWLNDSEWLREKITTS
GKHPQQPLVIVVGTSLAVYPFASLPEEIPRKVKRVLCNLETVGDFKANKRPTDLIVHQYSDEFAEQLVEE
LGWQEDFEKILTAQGGMGDNSKEQLLEIVHDLENLSLDQSEHESADKKDKKLQRLNGHDSDEDGASNSSS
SQKAAKE
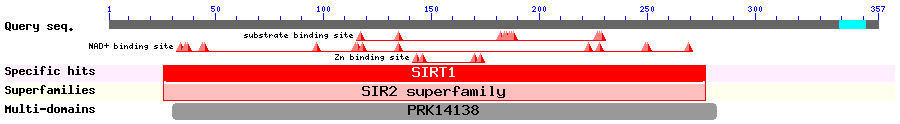
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 997877887899999999872999829998158744579899987899862234688999
Pred: CCCCCCCCCCCHHHHHHHHHCCCCCEEEEEECCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: MSVSTASTEMSVRKIAAHMKSNPNAKVIFMVGAGISTSCGIPDFRSPGTGLYHNLARLKL
10 20 30 40 50 60
Conf: 995532223443559933899876647999899923589984313685000134551247
Pred: CCCCCCCCCCCCCCCCHHHHHHHHHHCCCCCCCCHHHHHHHHHHCCCCCCCCCCCCCHHH
AA: PYPEAVFDVDFFQSDPLPFYTLAKELYPGNFRPSKFHYLLKLFQDKDVLKRVYTQNIDTL
70 80 90 100 110 120
Conf: 764199998566404642343234469657927899888459999988889999832588
Pred: HHHCCCCCCCEEECCCCCCCCCCCCCCCCCCHHHHHHHHHCCCCCCCCCCCCCCCCCCCC
AA: ERQAGVKDDLIIEAHGSFAHCHCIGCGKVYPPQVFKSKLAEHPIKDFVKCDVCGELVKPA
130 140 150 160 170 180
Conf: 002299991547885654357777654068999998989994645223436667898999
Pred: EEEECCCCCHHHHHHHHHHHHHHHHHHHCCCCCCCCCEEEEECCCCCCCCCCCCCCCCCC
AA: IVFFGEDLPDSFSETWLNDSEWLREKITTSGKHPQQPLVIVVGTSLAVYPFASLPEEIPR
190 200 210 220 230 240
Conf: 997688706546888888897661661558999999999909999999999971699999
Pred: CCCEEEEECCCCCCCCCCCCCCCEEECCCHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCC
AA: KVKRVLCNLETVGDFKANKRPTDLIVHQYSDEFAEQLVEELGWQEDFEKILTAQGGMGDN
250 260 270 280 290 300
Conf: 335665420023357867543333430134554306999998899999973000149
Pred: CHHHHHHHHCCCCCCCCCCCCCCCCCCHHHHHHHHCCCCCCCCCCCCCCCCCHHCCC
AA: SKEQLLEIVHDLENLSLDQSEHESADKKDKKLQRLNGHDSDEDGASNSSSSQKAAKE
310 320 330 340 350
Show Conservation Score by AL2CO
1 M -1.000 *
2 S -1.000 *
3 V -1.000 *
4 S -1.000 *
5 T -1.000 *
6 A -1.000 *
7 S -1.000 *
8 T -1.000 *
9 E -1.000 *
10 M -0.985
11 S -0.109
12 V 0.371
13 R -0.503
14 K -0.645
15 I 0.520
16 A -0.328
17 A -0.556
18 H -0.772
19 M 0.579
20 K -0.449
21 S -0.576
22 N -0.756
23 P -0.655
24 N -0.865
25 A -0.513
26 K 0.293
27 V 1.029
28 I -0.005
29 F 0.111
30 M 0.483
31 V -0.176
32 G 1.945
33 A 1.753
34 G 1.769
35 I 0.579
36 S 2.833
37 T 1.101
38 S 0.269
39 C 1.015
40 G 1.139
41 I 1.597
42 P 2.109
43 D 2.113
44 F 2.444
45 R 2.833
46 S 1.497
47 P 0.256
48 G -0.410
49 T -0.356
50 G 2.833
51 L 0.630
52 Y 1.620
53 H -0.322
54 N -0.020
55 L 0.177
56 A -0.424
57 R -0.683
58 L -0.212
59 K -0.176
60 L 0.992
61 P 0.125
62 Y -0.390
63 P 1.396
64 E 0.101
65 A 0.159
66 V 0.823
67 F 1.696
68 D 0.105
69 V 0.015
70 D -0.517
71 F 0.082
72 F 1.558
73 Q -0.928
74 S -0.856
75 D -0.115
76 P 1.627
77 L -0.851
78 P 0.181
79 F 2.107
80 Y 0.731
81 T -0.823
82 L 0.302
83 A -0.171
84 K -0.567
85 E -0.639
86 L 0.452
87 Y -0.627
88 P -0.142
89 G -0.511
90 N -0.860
91 F -0.517
92 R -0.954
93 P 0.448
94 S 0.966
95 K -0.804
96 F -0.306
97 H 2.484
98 Y -0.795
99 L 1.131
100 L 0.178
101 K -0.128
102 L -0.562
103 F 0.336
104 Q -0.628
105 D -0.515
106 K -0.226
107 D -0.168
108 V -0.553
109 L 1.513
110 K -0.591
111 R -0.192
112 V -0.242
113 Y 0.492
114 T 1.476
115 Q 2.131
116 N 2.097
117 I 1.280
118 D 2.438
119 T -0.111
120 L 1.624
121 E 1.391
122 R -0.721
123 Q -0.727
124 A 0.657
125 G 0.465
126 V 0.634
127 K -0.529
128 D -0.672
129 D -0.323
130 L -0.851
131 I 0.347
132 I 0.424
133 E -0.403
134 A 0.173
135 H 2.103
136 G 2.009
137 S 0.119
138 F -0.029
139 A -0.730
140 H -0.736
141 C -0.028
142 H -0.324
143 C 2.104
144 I -0.545
145 G -0.758
146 C 2.084
147 G -0.577
148 K -0.861
149 V -0.937
150 Y -0.655
151 P -0.693
152 P -1.082
153 Q -0.789
154 V -1.025
155 F -0.417
156 K -0.806
157 S -0.653
158 K -0.989
159 L 0.047
160 A -0.891
161 E -0.769
162 H -0.893
163 P -0.937
164 I -0.603
165 K -0.755
166 D -0.992
167 F -0.528
168 V -0.708
169 K -0.996
170 C -0.776
171 D -0.916
172 V -1.053
173 C -0.819
174 G -0.983
175 E 0.101
176 L -0.047
177 V 0.760
178 K 1.576
179 P 1.606
180 A -0.026
181 I 1.239
182 V 0.879
183 F 1.345
184 F 1.254
185 G 0.715
186 E 1.651
187 D -0.496
188 L 1.271
189 P 1.130
190 D -0.861
191 S -0.720
192 F 0.746
193 S -0.961
194 E -0.700
195 T -0.843
196 W -0.717
197 L -0.993
198 N -0.831
199 D 0.199
200 S -1.000 *
201 E -1.000 *
202 W -1.000 *
203 L -1.000 *
204 R -1.000 *
205 E -1.000 *
206 K -1.000 *
207 I -1.000 *
208 T -1.000 *
209 T -1.000 *
210 S -1.000 *
211 G -1.000 *
212 K -1.000 *
213 H -0.614
214 P -0.942
215 Q -0.793
216 Q -0.390
217 P 0.404
218 L 0.674
219 V 0.250
220 I 0.824
221 V 1.048
222 V 0.167
223 G 2.833
224 T 1.982
225 S 1.686
226 L 2.219
227 A -0.437
228 V 1.643
229 Y -0.528
230 P 2.833
231 F 0.700
232 A 0.797
233 S -0.549
234 L 0.802
235 P -0.017
236 E -0.954
237 E -1.031
238 I -0.246
239 P -0.739
240 R -1.034
241 K -0.704
242 V -0.384
243 K -0.343
244 R 0.500
245 V 0.269
246 L 0.894
247 C 0.467
248 N 2.077
249 L -0.151
250 E -0.155
251 T -0.860
252 V -0.092
253 G -0.740
254 D -0.767
255 F -0.786
256 K -0.770
257 A -1.151
258 N -0.943
259 K -0.897
260 R -0.732
261 P -1.007
262 T -0.689
263 D 2.022
264 L -0.152
265 I -0.955
266 V -0.675
267 H -0.688
268 Q -0.148
269 Y -0.661
270 S 0.079
271 D -0.212
272 E -0.836
273 F -0.683
274 A 0.139
275 E -1.046
276 Q -0.844
277 L 0.870
278 V -0.100
279 E -0.535
280 E -0.812
281 L 0.448
282 G 0.073
283 W 0.021
284 Q -0.927
285 E -0.928
286 D -0.423
287 F -0.472
288 E -0.736
289 K -0.728
290 I -0.637
291 L -1.008
292 T -0.975
293 A -0.935
294 Q -1.003
295 G -1.111
296 G -0.922
297 M -0.966
298 G -1.091
299 D -0.923
300 N -1.043
301 S -1.055
302 K -0.993
303 E -0.946
304 Q -0.872
305 L -0.915
306 L -1.038
307 E -0.889
308 I -0.980
309 V -1.034
310 H -1.059
311 D -0.853
312 L -1.062
313 E -0.942
314 N -0.981
315 L -1.000 *
316 S -1.000 *
317 L -1.000 *
318 D -1.000 *
319 Q -1.000 *
320 S -1.000 *
321 E -1.000 *
322 H -1.000 *
323 E -1.000 *
324 S -1.000 *
325 A -1.000 *
326 D -1.000 *
327 K -1.000 *
328 K -1.000 *
329 D -1.000 *
330 K -1.000 *
331 K -1.000 *
332 L -1.000 *
333 Q -1.000 *
334 R -1.000 *
335 L -1.000 *
336 N -1.000 *
337 G -1.000 *
338 H -1.000 *
339 D -1.000 *
340 S -1.000 *
341 D -1.000 *
342 E -1.000 *
343 D -1.000 *
344 G -1.000 *
345 A -1.000 *
346 S -1.000 *
347 N -1.000 *
348 S -1.000 *
349 S -1.000 *
350 S -1.000 *
351 S -1.000 *
352 Q -1.000 *
353 K -1.000 *
354 A -1.000 *
355 A -1.000 *
356 K -1.000 *
357 E -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 212.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MSVSTASTEM SVRKIAAHMK SNPNAKVIFM VGAGISTSCG IPDFRSPGTG LYHNLARLKL
70 80 90 100 110 120
PYPEAVFDVD FFQSDPLPFY TLAKELYPGN FRPSKFHYLL KLFQDKDVLK RVYTQNIDTL
130 140 150 160 170 180
ERQAGVKDDL IIEAHGSFAH CHCIGCGKVY PPQVFKSKLA EHPIKDFVKC DVCGELVKPA
190 200 210 220 230 240
IVFFGEDLPD SFSETWLNDS EWLREKITTS GKHPQQPLVI VVGTSLAVYP FASLPEEIPR
250 260 270 280 290 300
KVKRVLCNLE TVGDFKANKR PTDLIVHQYS DEFAEQLVEE LGWQEDFEKI LTAQGGMGDN
310 320 330 340 350
SKEQLLEIVH DLENLSLDQS EHESADKKDK KLQRLNGHDS DEDGASNSSS SQKAAKE
Hst2 is a a homologue of SIR2 2 protein. It deacetylates histones in vivo and participates in the transcriptional repression of FLO10 through direct binding of its promoter. Despite its nuclear function, it is localized to the cytoplasm, which is a result of rapid nuclear export. Hst2 shared a 40% sequence identity to SIRT2 (NES ID 199). However, the two NESs don't align.
[1]. "Nuclear export modulates the cytoplasmic Sir2 homologue Hst2."
Wilson JM, Le VQ, Zimmerman C, Marmorstein R, Pillus L (2006)
EMBO Rep,
7:1247-51
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
