Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform
UniProt
helix followed by a short loop and then β-strand as part of an extend β-sheet
Show FASTA Format
>gi|54038809|sp|P67775.1|PP2AA_HUMAN RecName: Full=Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform; Short=PP2A-alpha; AltName: Full=Replication protein C; Short=RP-C
MDEKVFTKELDQWIEQLNECKQLSESQVKSLCEKAKEILTKESNVQEVRCPVTVCGDVHGQFHDLMELFR
IGGKSPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYRERITILRGNHESRQITQVYGFYDECLRKYGNA
NVWKYFTDLFDYLPLTALVDGQIFCLHGGLSPSIDTLDHIRALDRLQEVPHEGPMCDLLWSDPDDRGGWG
ISPRGAGYTFGQDISETFNHANGLTLVSRAHQLVMEGYNWCHDRNVVTIFSAPNYCYRCGNQAAIMELDD
TLKYSFLQFDPAPRRGEPHVTRRTPDYFL
Show Domain Info by CDD
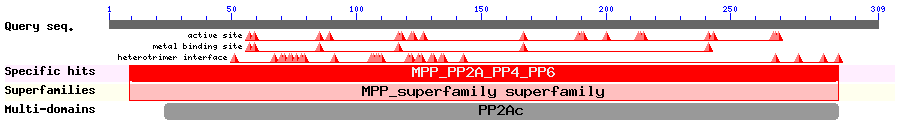
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 985337135999999983088999789999999999986116892232287445035541
Pred: CCCCCCCHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHCCCCEEECCCEEEECCCCC
AA: MDEKVFTKELDQWIEQLNECKQLSESQVKSLCEKAKEILTKESNVQEVRCPVTVCGDVHG
10 20 30 40 50 60
Conf: 245389998822999996300132334566523548888865520268836784267764
Pred: HHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHCCCCCEEEECCCCCC
AA: QFHDLMELFRIGGKSPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYRERITILRGNHES
70 80 90 100 110 120
Conf: 221100003577753108820254642102555302112182886348888884434676
Pred: CCCCEEEHHHHHHHHHCCCCHHHHHHHHCCCCCCCCEEECCCEEEECCCCCCCCCCHHHH
AA: RQITQVYGFYDECLRKYGNANVWKYFTDLFDYLPLTALVDGQIFCLHGGLSPSIDTLDHI
130 140 150 160 170 180
Conf: 520124457999984234678999988878898887532350046764311796010033
Pred: HHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHCCCCCEEHHH
AA: RALDRLQEVPHEGPMCDLLWSDPDDRGGWGISPRGAGYTFGQDISETFNHANGLTLVSRA
190 200 210 220 230 240
Conf: 343321544422582899941698500026403454312235832544238999999988
Pred: HHHHHCCCCCCCCCCEEEEEECCCCCCCCCCCEEEEEECCCCCCEEEEECCCCCCCCCCC
AA: HQLVMEGYNWCHDRNVVTIFSAPNYCYRCGNQAAIMELDDTLKYSFLQFDPAPRRGEPHV
250 260 270 280 290 300
Conf: 889999999
Pred: CCCCCCCCC
AA: TRRTPDYFL
Show Conservation Score by AL2CO
1 M -1.000 *
2 D -1.000 *
3 E -1.000 *
4 K -1.000 *
5 V -1.000 *
6 F -1.000 *
7 T -1.000 *
8 K -1.000 *
9 E -0.734
10 L 0.088
11 D 0.061
12 Q -1.135
13 W -1.226
14 I -0.331
15 E -0.929
16 Q -1.274
17 L -0.303
18 N -1.175
19 E -0.958
20 C -1.098
21 K -0.763
22 Q -1.346
23 L 0.378
24 S -1.076
25 E 0.116
26 S -1.316
27 Q -0.720
28 V -0.132
29 K -1.223
30 S -1.375
31 L 0.465
32 C 0.418
33 E -1.338
34 K -1.198
35 A -0.386
36 K -0.713
37 E -0.777
38 I -0.379
39 L 0.336
40 T -1.004
41 K -1.008
42 E 0.774
43 S -0.862
44 N -0.126
45 V 0.275
46 Q -0.986
47 E -1.405
48 V 0.318
49 R -1.195
50 C -0.660
51 P 2.035
52 V 0.189
53 T -0.412
54 V 0.662
55 C 0.077
56 G 2.035
57 D 1.700
58 V 0.671
59 H 2.035
60 G 1.583
61 Q 2.035
62 F 0.445
63 H -1.105
64 D 2.035
65 L 0.677
66 M -0.812
67 E -0.073
68 L 0.465
69 F 0.625
70 R -0.929
71 I -1.025
72 G 0.243
73 G 0.182
74 K -1.376
75 S -0.702
76 P -0.407
77 D -1.116
78 T -0.937
79 N -0.502
80 Y 0.531
81 L 0.466
82 F 1.328
83 M 0.104
84 G 1.096
85 D 0.926
86 Y 0.646
87 V 1.057
88 D 1.310
89 R 1.397
90 G 1.695
91 Y -0.711
92 Y -0.409
93 S 0.789
94 V 0.268
95 E 1.142
96 T 0.147
97 V -0.343
98 T -0.461
99 L -0.508
100 L 0.284
101 V -0.315
102 A -0.302
103 L 0.247
104 K 1.329
105 V 0.207
106 R 0.296
107 Y 0.544
108 R 0.292
109 E -0.832
110 R -0.118
111 I 0.238
112 T -0.868
113 I 0.286
114 L 0.464
115 R 2.035
116 G 2.035
117 N 2.035
118 H 2.035
119 E 2.035
120 S -0.010
121 R 0.455
122 Q -0.606
123 I 0.252
124 T 0.688
125 Q -0.303
126 V 0.026
127 Y 1.699
128 G 1.408
129 F 1.327
130 Y 0.871
131 D -0.050
132 E 1.700
133 C 0.376
134 L -1.057
135 R -0.098
136 K 0.487
137 Y 0.277
138 G -0.127
139 N -0.741
140 A -1.018
141 N -1.092
142 V -0.239
143 W 1.700
144 K -0.441
145 Y -1.271
146 F 0.552
147 T -0.117
148 D -0.610
149 L -0.307
150 F 2.035
151 D 0.453
152 Y -0.513
153 L 0.627
154 P -0.211
155 L -0.040
156 T -0.059
157 A 0.277
158 L -0.467
159 V 0.588
160 D -0.239
161 G -0.547
162 Q -0.828
163 I -0.529
164 F 0.444
165 C 0.369
166 L -0.477
167 H 2.035
168 G 1.381
169 G 2.035
170 L 1.202
171 S 2.035
172 P 1.700
173 S -1.081
174 I -0.333
175 D -1.162
176 T -1.029
177 L 0.155
178 D -0.255
179 H -0.864
180 I 0.204
181 R -0.836
182 A -1.333
183 L 0.107
184 D -1.184
185 R 1.698
186 L -1.283
187 Q -0.918
188 E 0.101
189 V 0.032
190 P 0.888
191 H -0.674
192 E -0.990
193 G 1.381
194 P -0.450
195 M -0.130
196 C 0.002
197 D 0.875
198 L -0.319
199 L 0.009
200 W 0.874
201 S 0.851
202 D 1.107
203 P 1.607
204 D -0.912
205 D -1.179
206 R -1.134
207 G -1.346
208 G -1.177
209 W 0.757
210 G -0.938
211 I -1.287
212 S 1.033
213 P -0.546
214 R 2.035
215 G 1.692
216 A 0.237
217 G 0.838
218 Y -0.269
219 T -0.494
220 F 0.660
221 G 0.820
222 Q -1.230
223 D -0.961
224 I -0.727
225 S -0.182
226 E -1.221
227 T -1.042
228 F 0.233
229 N -0.736
230 H -1.196
231 A -1.275
232 N 0.163
233 G -0.675
234 L -0.142
235 T -0.976
236 L -0.823
237 V 0.525
238 S -0.371
239 R 1.694
240 A 0.736
241 H 2.035
242 Q 1.322
243 L 0.706
244 V 0.195
245 M -0.549
246 E 0.063
247 G 2.035
248 Y -0.035
249 N -0.781
250 W -1.198
251 C -1.035
252 H -0.854
253 D -0.678
254 R -0.822
255 N -1.050
256 V 0.047
257 V -0.029
258 T 1.638
259 I 0.784
260 F 1.211
261 S 1.309
262 A 1.407
263 P 2.035
264 N 2.035
265 Y 2.035
266 C 0.630
267 Y 0.600
268 R 1.063
269 C -0.052
270 G -0.031
271 N 2.035
272 Q -0.983
273 A 1.042
274 A 0.320
275 I 0.314
276 M 0.274
277 E -1.011
278 L -0.029
279 D -0.732
280 D -0.642
281 T -1.313
282 L -1.231
283 K -1.113
284 Y -1.170
285 S -1.095
286 F -0.607
287 L -1.041
288 Q -0.848
289 F 0.186
290 D -0.964
291 P -0.570
292 A -0.686
293 P -0.786
294 R -1.084
295 R -1.129
296 G -1.299
297 E -1.204
298 P -1.356
299 H -1.369
300 V -1.290
301 T -1.368
302 R -1.036
303 R -1.116
304 T -1.145
305 P -0.747
306 D -0.457
307 Y 1.573
308 F 1.069
309 L 1.162
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 213.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MDEKVFTKEL DQWIEQLNEC KQLSESQVKS LCEKAKEILT KESNVQEVRC PVTVCGDVHG
70 80 90 100 110 120
QFHDLMELFR IGGKSPDTNY LFMGDYVDRG YYSVETVTLL VALKVRYRER ITILRGNHES
130 140 150 160 170 180
RQITQVYGFY DECLRKYGNA NVWKYFTDLF DYLPLTALVD GQIFCLHGGL SPSIDTLDHI
190 200 210 220 230 240
RALDRLQEVP HEGPMCDLLW SDPDDRGGWG ISPRGAGYTF GQDISETFNH ANGLTLVSRA
250 260 270 280 290 300
HQLVMEGYNW CHDRNVVTIF SAPNYCYRCG NQAAIMELDD TLKYSFLQFD PAPRRGEPHV
TRRTPDYFL
2IAE (X-Ray,3.50 Å resolution)
3C5W (X-Ray,2.80 Å resolution)
3FGA (X-Ray,2.70 Å resolution)
Protein phosphatase 2A is a holoenzyme consisting of three subunits. Tsuchiya et al. showed that LMB induces nuclear accumulation of PP2A. The sustained dephosphorylation activity of PP2A is involved in the inhibition of cyclin D1 expression by LMB, which induces G1 arrest in NIH3T2 cells. Note that an NES was also identified in the regulatory subunit of PP2A (NES ID 157).
[1]. "Involvement of protein phosphatase 2A nuclear accumulation and subsequent inactivation of activator protein-1 in leptomycin B-inhibited cyclin D1 expression."
Tsuchiya A, Tashiro E, Yoshida M, Imoto M (2007)
Oncogene,
26:1522-32
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
