Summary for HSCARG (NES ID: 225)
Full Name
NmrA-like family domain-containing protein 1
UniProt Alternative Names
None
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
Secondary Structure of Export Signal
short α-helix flanked by loops
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|74734255|sp|Q9HBL8.1|NMRL1_HUMAN RecName: Full=NmrA-like family domain-containing protein 1
MVDKKLVVVFGGTGAQGGSVARTLLEDGTFKVRVVTRNPRKKAAKELRLQGAEVVQGDQDDQVIMELALN
GAYATFIVTNYWESCSQEQEVKQGKLLADLARRLGLHYVVYSGLENIKKLTAGRLAAAHFDGKGEVEEYF
RDIGVPMTSVRLPCYFENLLSHFLPQKAPDGKSYLLSLPTGDVPMDGMSVSDLGPVVLSLLKMPEKYVGQ
NIGLSTCRHTAEEYAALLTKHTRKVVHDAKMTPEDYEKLGFPGARDLANMFRFYALRPDRDIELTLRLNP
KALTLDQWLEQHKGDFNLL
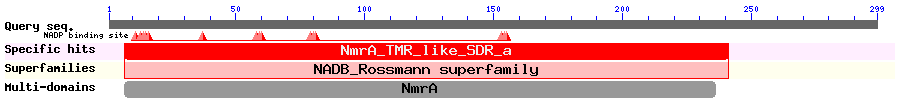
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 999986999777993218999999848997189955899959899998799939980499
Pred: CCCCCEEEEECCCCCCHHHHHHHHHHCCCCEEEEECCCCCCHHHHHHHHCCCEEEECCCC
AA: MVDKKLVVVFGGTGAQGGSVARTLLEDGTFKVRVVTRNPRKKAAKELRLQGAEVVQGDQD
10 20 30 40 50 60
Conf: 998899987389499980899999992489999999999999869998998688897523
Pred: CHHHHHHHHCCCCEEEECCCCCCCCCHHHHHHHHHHHHHHHHHCCCCEEEECCCCCCCCC
AA: DQVIMELALNGAYATFIVTNYWESCSQEQEVKQGKLLADLARRLGLHYVVYSGLENIKKL
70 80 90 100 110 120
Conf: 699423587677899999998649993897347645531106899636999647852899
Pred: CCCCCCCCCCCCHHHHHHHHHHCCCCEEEEECCHHHHHHHCCCCCCCCCCCCEEEECCCC
AA: TAGRLAAAHFDGKGEVEEYFRDIGVPMTSVRLPCYFENLLSHFLPQKAPDGKSYLLSLPT
130 140 150 160 170 180
Conf: 999887416208799999995099856991899861678999999999976199320211
Pred: CCCCCCCCCCCCHHHHHHHHHHCCCCCCCCEEEEECCCCCHHHHHHHHHHHHCCCCCCCC
AA: GDVPMDGMSVSDLGPVVLSLLKMPEKYVGQNIGLSTCRHTAEEYAALLTKHTRKVVHDAK
190 200 210 220 230 240
Conf: 69888973299949999999998212999998889764999999999998650135589
Pred: CCHHHHHHCCCCCHHHHHHHHHHHCCCCCCCHHHHHHHCCCCCCHHHHHHHCCCCCCCC
AA: MTPEDYEKLGFPGARDLANMFRFYALRPDRDIELTLRLNPKALTLDQWLEQHKGDFNLL
250 260 270 280 290
Show Conservation Score by AL2CO
1 M 4.731
2 V 0.107
3 D -0.212
4 K -0.299
5 K 0.423
6 L -0.304
7 V 1.532
8 V 0.849
9 V 3.441
10 F 0.321
11 G 2.840
12 G 1.788
13 T 2.181
14 G 3.022
15 A -0.538
16 Q 1.619
17 G 4.312
18 G 0.256
19 S 1.099
20 V 1.430
21 A 1.179
22 R -0.269
23 T -0.478
24 L 1.515
25 L 0.349
26 E -0.070
27 D -0.319
28 G 0.252
29 T -0.728
30 F 0.175
31 K -0.515
32 V 2.194
33 R 1.018
34 V 1.567
35 V 1.010
36 T 1.201
37 R 2.550
38 N 0.588
39 P 0.250
40 R -0.333
41 K 0.311
42 K -0.336
43 A -0.112
44 A 1.707
45 K 0.103
46 E -0.465
47 L 2.753
48 R -0.555
49 L -0.466
50 Q -0.507
51 G 1.416
52 A 1.247
53 E 0.059
54 V 0.866
55 V 0.140
56 Q -0.566
57 G 0.872
58 D 0.460
59 Q -0.341
60 D 0.038
61 D -0.233
62 Q -0.789
63 V -0.521
64 I -0.245
65 M 1.168
66 E -0.815
67 L -0.564
68 A 0.886
69 L 0.482
70 N -0.353
71 G 0.958
72 A 0.690
73 Y -0.294
74 A 0.114
75 T 0.715
76 F 1.829
77 I 0.391
78 V 0.671
79 T 0.415
80 N -0.208
81 Y -0.392
82 W -1.028
83 E -0.755
84 S -1.065
85 C -1.192
86 S -0.343
87 Q -1.043
88 E -0.956
89 Q -0.915
90 E 3.067
91 V -0.760
92 K -1.053
93 Q -0.716
94 G 1.269
95 K -0.526
96 L -0.591
97 L 0.426
98 A 0.302
99 D 0.285
100 L 0.350
101 A 1.184
102 R -0.368
103 R -0.707
104 L -0.687
105 G 0.240
106 L 1.493
107 H -0.513
108 Y 0.636
109 V 0.864
110 V 2.503
111 Y 0.925
112 S 1.066
113 G 1.507
114 L 0.253
115 E -0.780
116 N 0.046
117 I -0.229
118 K -0.941
119 K -0.436
120 L -1.140
121 T -0.352
122 A -0.151
123 G 0.032
124 R -0.656
125 L -0.198
126 A -1.074
127 A -0.106
128 A -0.896
129 H -0.192
130 F 0.463
131 D 0.228
132 G -0.464
133 K 3.644
134 G -0.718
135 E -0.678
136 V 1.211
137 E 0.503
138 E -0.320
139 Y 0.133
140 F 0.758
141 R -0.649
142 D -0.426
143 I -0.499
144 G -0.249
145 V 0.076
146 P -0.526
147 M -0.669
148 T 1.028
149 S -0.304
150 V 0.712
151 R -0.851
152 L -0.115
153 P 0.659
154 C -0.501
155 Y 2.234
156 F 0.180
157 E 0.424
158 N 0.425
159 L 0.247
160 L -1.038
161 S -0.774
162 H -1.112
163 F -0.809
164 L -0.890
165 P -0.497
166 Q -1.008
167 K -0.991
168 A -1.019
169 P -0.689
170 D -0.575
171 G -0.820
172 K -1.019
173 S -1.166
174 Y -1.032
175 L -1.064
176 L -0.789
177 S -1.054
178 L -0.912
179 P -0.453
180 T -1.072
181 G -0.845
182 D -0.758
183 V -0.901
184 P -0.527
185 M -0.591
186 D -0.723
187 G -0.781
188 M -0.072
189 S 0.155
190 V -0.246
191 S -0.465
192 D 1.659
193 L 0.106
194 G 3.671
195 P -0.942
196 V 0.181
197 V 0.813
198 L -0.619
199 S -1.005
200 L 0.679
201 L 1.212
202 K -0.403
203 M -0.392
204 P -0.259
205 E -0.264
206 K -0.480
207 Y -0.270
208 V -0.913
209 G 0.835
210 Q -0.440
211 N -0.756
212 I 0.596
213 G -1.101
214 L 0.201
215 S 0.122
216 T 0.165
217 C -0.221
218 R -1.092
219 H -0.527
220 T 0.630
221 A -0.637
222 E -0.709
223 E 0.013
224 Y 0.152
225 A -0.190
226 A -0.230
227 L -0.441
228 L 0.796
229 T -0.151
230 K -0.204
231 H -0.617
232 T -0.321
233 R -0.155
234 K -0.809
235 V -0.548
236 V -0.216
237 H -0.974
238 D -0.606
239 A -0.969
240 K -0.797
241 M -0.401
242 T -0.476
243 P -1.101
244 E -0.239
245 D -0.800
246 Y -0.103
247 E -0.879
248 K -0.704
249 L -1.025
250 G -1.035
251 F -0.964
252 P -0.828
253 G -0.951
254 A -0.420
255 R -0.716
256 D -0.531
257 L -0.266
258 A -0.937
259 N -0.853
260 M -0.073
261 F -0.524
262 R -0.502
263 F -0.194
264 Y -0.331
265 A -0.884
266 L -1.183
267 R -1.165
268 P -0.905
269 D -0.692
270 R -0.340
271 D 0.155
272 I -0.590
273 E -0.282
274 L -0.797
275 T 0.253
276 L -0.659
277 R -0.344
278 L -0.508
279 N -0.623
280 P 0.027
281 K -0.457
282 A -0.451
283 L -0.427
284 T 0.764
285 L 0.843
286 D 0.049
287 Q 0.083
288 W 2.166
289 L 0.928
290 E -0.495
291 Q -0.232
292 H 0.062
293 K -0.342
294 G -0.434
295 D -1.000 *
296 F -1.000 *
297 N -1.000 *
298 L -1.000 *
299 L -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 225.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MVDKKLVVVF GGTGAQGGSV ARTLLEDGTF KVRVVTRNPR KKAAKELRLQ GAEVVQGDQD
70 80 90 100 110 120
DQVIMELALN GAYATFIVTN YWESCSQEQE VKQGKLLADL ARRLGLHYVV YSGLENIKKL
130 140 150 160 170 180
TAGRLAAAHF DGKGEVEEYF RDIGVPMTSV RLPCYFENLL SHFLPQKAPD GKSYLLSLPT
190 200 210 220 230 240
GDVPMDGMSV SDLGPVVLSL LKMPEKYVGQ NIGLSTCRHT AEEYAALLTK HTRKVVHDAK
250 260 270 280 290
MTPEDYEKLG FPGARDLANM FRFYALRPDR DIELTLRLNP KALTLDQWLE QHKGDFNLL
3D Structures in PDB
2EXX (X-Ray,2.40 Å resolution)
3DXF (X-Ray,2.20 Å resolution)
3E5M (X-Ray,2.70 Å resolution)
Comments
HSCARG is an inhibitor of nuclear factor-κB. HSCARG blocks basal and stimulus-coupled NF-κB by regulating the ubiquitination of the NF-κB subunit RelA. HSCARG also interacts with IκB kinase complex, inhibits the phosphorylation of IKKβ, blocks the degradation of IκBα and further suppresses NF-κB activity. Zhang et al. showed that HSCARG contains one NES and mutation of the NES decreases the interaction between HSCARG and IKKβ , thus weakened the inhibition of NF-κB.
References
[1]. "A CRM1-Dependent Nuclear Export Signal Controls Nucleocytoplasmic Translocation of HSCARG, Which Regulates NF-kappaB Activity."
Zhang M, Hu B, Li T, Peng Y, Guan J, Lai S, Zheng X (2012)
Traffic,
13:790-9
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
