Mesoderm induction early response protein 1
UniProt
Show FASTA Format
>gi|225735610|ref|NP_001071170.2| mesoderm induction early response protein 1 isoform d precursor [Homo sapiens]
MFMFNWFTDCLWTLFLSNYQPSVESSSPGGSATSDDHEFDPSADMLVHDFDDERTLEEEEMMEGETNFSS
EIEDLAREGDMPIHELLSLYGYGSTVRLPEEDEEEEEEEEEGEDDEDADNDDNSGCSGENKEENIKDSSG
QEDETQSSNDDPSQSVASQDAQEIIRPRRCKYFDTNSEVEEESEEDEDYIPSEDWKKEIMVGSMFQAEIP
VGICRYKENEKVYENDDQLLWDPEYLPEDKVIIFLKDASRRTGDEKGVEAIPEGSHIKDNEQALYELVKC
NFDTEEALRRLRFNVKAAREELSVWTEEECRNFEQGLKAYGKDFHLIQANKVRTRSVGECVAFYYMWKKS
ERYDFFAQQTRFGKKKYNLHPGVTDYMDRLLDESESAASSRAPSPPPTASNSSNSQSEKEDGTVSTANQN
GVSSNGPGILQMLLPVHFSAISSRANAFLK
Show Domain Info by CDD
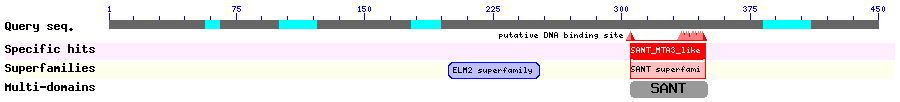
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 965303477887522777999987889998999999889920110026886100002764
Pred: CCCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHCCCCCCCCCCHHHH
AA: MFMFNWFTDCLWTLFLSNYQPSVESSSPGGSATSDDHEFDPSADMLVHDFDDERTLEEEE
10 20 30 40 50 60
Conf: 135999973236788750999999999861899999999876402444202888867788
Pred: HCCCCCCCCHHHHHHHHHCCCCHHHHHHHHCCCCCCCCCCCCCHHHHHHHCCCCCCCCCC
AA: MMEGETNFSSEIEDLAREGDMPIHELLSLYGYGSTVRLPEEDEEEEEEEEEGEDDEDADN
70 80 90 100 110 120
Conf: 899999987621123588999775567899999876451002101864346679998865
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCCCCCCC
AA: DDNSGCSGENKEENIKDSSGQEDETQSSNDDPSQSVASQDAQEIIRPRRCKYFDTNSEVE
130 140 150 160 170 180
Conf: 666555679996224200146775111268756688755223686565454899999689
Pred: CCCCCCCCCCCCHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHH
AA: EESEEDEDYIPSEDWKKEIMVGSMFQAEIPVGICRYKENEKVYENDDQLLWDPEYLPEDK
190 200 210 220 230 240
Conf: 999999998740776586678988766891999999972699989999972488425763
Pred: HHHHHHHHHHHCCCCCCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHCCCCCCCCC
AA: VIIFLKDASRRTGDEKGVEAIPEGSHIKDNEQALYELVKCNFDTEEALRRLRFNVKAARE
250 260 270 280 290 300
Conf: 357779999985999999817523677730144445331102331012440247888876
Pred: CCCCCCHHHHHHHHHHHHHHCCCHHHHHHCCCCCCCCCCCEEEEECCCCCCHHHHHHHHH
AA: ELSVWTEEECRNFEQGLKAYGKDFHLIQANKVRTRSVGECVAFYYMWKKSERYDFFAQQT
310 320 330 340 350 360
Conf: 611343468999876101122311224799999999989999999998755655667899
Pred: HHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: RFGKKKYNLHPGVTDYMDRLLDESESAASSRAPSPPPTASNSSNSQSEKEDGTVSTANQN
370 380 390 400 410 420
Conf: 988899997543478898789853222469
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: GVSSNGPGILQMLLPVHFSAISSRANAFLK
430 440 450
Show Conservation Score by AL2CO
1 M -1.000 *
2 F -1.000 *
3 M -1.000 *
4 F -1.000 *
5 N -1.000 *
6 W -1.000 *
7 F -1.000 *
8 T -1.000 *
9 D -1.000 *
10 C -1.000 *
11 L -1.000 *
12 W -1.000 *
13 T -1.000 *
14 L -1.000 *
15 F -1.000 *
16 L -1.000 *
17 S -1.000 *
18 N -1.000 *
19 Y -1.000 *
20 Q -1.000 *
21 P -1.000 *
22 S -1.000 *
23 V -1.000 *
24 E -1.000 *
25 S -1.000 *
26 S -1.000 *
27 S -1.000 *
28 P -1.000 *
29 G -1.000 *
30 G -1.000 *
31 S -1.000 *
32 A -1.000 *
33 T -1.000 *
34 S -1.000 *
35 D -1.000 *
36 D -1.000 *
37 H -1.000 *
38 E -1.000 *
39 F -1.000 *
40 D -1.000 *
41 P -1.000 *
42 S -1.000 *
43 A -1.000 *
44 D -1.000 *
45 M -1.000 *
46 L -1.000 *
47 V -1.000 *
48 H -1.000 *
49 D -1.000 *
50 F -1.000 *
51 D -1.000 *
52 D -1.000 *
53 E -1.000 *
54 R -1.000 *
55 T -1.000 *
56 L -1.000 *
57 E -1.000 *
58 E -1.000 *
59 E -1.000 *
60 E -1.000 *
61 M -1.000 *
62 M -1.000 *
63 E -1.000 *
64 G -1.000 *
65 E -1.000 *
66 T -1.000 *
67 N -1.000 *
68 F -1.000 *
69 S -1.000 *
70 S -1.000 *
71 E -1.000 *
72 I 1.118
73 E 0.437
74 D -0.086
75 L 1.483
76 A 0.065
77 R 0.941
78 E 0.771
79 G 1.032
80 D 1.385
81 M 0.904
82 P 1.898
83 I 0.892
84 H 1.093
85 E 1.270
86 L 1.642
87 L 0.782
88 S 0.226
89 L 0.804
90 Y 1.319
91 G 0.061
92 Y 0.242
93 G -1.000 *
94 S -1.000 *
95 T -1.000 *
96 V -1.000 *
97 R -1.000 *
98 L -1.000 *
99 P -1.000 *
100 E -0.514
101 E -0.242
102 D -0.297
103 E 0.290
104 E -0.096
105 E -0.270
106 E -0.075
107 E -1.000 *
108 E -1.000 *
109 E -1.000 *
110 E -1.000 *
111 E -1.000 *
112 G -1.000 *
113 E -1.000 *
114 D -1.000 *
115 D -1.000 *
116 E -1.000 *
117 D -1.000 *
118 A -1.000 *
119 D -0.552
120 N -0.556
121 D -0.364
122 D -0.831
123 N -0.646
124 S -0.582
125 G -1.000 *
126 C -1.000 *
127 S -1.000 *
128 G -1.000 *
129 E -1.000 *
130 N -0.421
131 K -0.125
132 E -0.396
133 E 0.088
134 N -0.636
135 I -0.247
136 K -0.545
137 D -0.218
138 S -0.778
139 S -0.459
140 G -0.628
141 Q -0.690
142 E -0.174
143 D -0.098
144 E -0.379
145 T -0.348
146 Q -0.260
147 S -0.429
148 S -0.462
149 N -0.662
150 D -0.366
151 D -0.308
152 P -0.360
153 S -0.642
154 Q -0.476
155 S -0.746
156 V -0.580
157 A -0.540
158 S -0.584
159 Q -0.720
160 D -0.599
161 A -0.462
162 Q -0.531
163 E -0.655
164 I -0.471
165 I -0.555
166 R -0.487
167 P -0.509
168 R -0.748
169 R -0.554
170 C -0.781
171 K -0.730
172 Y -0.970
173 F -0.755
174 D -0.482
175 T -0.693
176 N -0.623
177 S -0.776
178 E -0.563
179 V -0.942
180 E -0.634
181 E -0.673
182 E -0.623
183 S -0.488
184 E -0.154
185 E -0.476
186 D -0.399
187 E -0.178
188 D -0.652
189 Y -0.556
190 I -0.737
191 P -0.073
192 S -0.676
193 E -0.312
194 D 0.195
195 W -0.886
196 K -0.416
197 K 0.443
198 E -0.515
199 I 0.167
200 M 0.703
201 V 0.468
202 G 0.660
203 S -0.496
204 M -0.160
205 F 0.950
206 Q 1.354
207 A 0.536
208 E -0.452
209 I 0.728
210 P 0.186
211 V -0.367
212 G -0.840
213 I -0.574
214 C -0.816
215 R -0.759
216 Y -0.914
217 K -0.647
218 E -0.994
219 N -0.801
220 E -0.785
221 K -0.435
222 V -0.723
223 Y -0.668
224 E -0.028
225 N -0.313
226 D -0.186
227 D 1.350
228 Q -0.192
229 L 0.848
230 L 2.095
231 W 3.925
232 D -0.328
233 P 0.862
234 E -0.559
235 Y -0.848
236 L -0.203
237 P -0.356
238 E 0.313
239 D -0.731
240 K -0.169
241 V 0.622
242 I -0.095
243 I -0.089
244 F 1.230
245 L 0.615
246 K -0.673
247 D -0.385
248 A 0.381
249 S -0.424
250 R 0.121
251 R -0.293
252 T -0.564
253 G -0.637
254 D -0.592
255 E -0.460
256 K -0.865
257 G -0.458
258 V -0.526
259 E -0.852
260 A -0.740
261 I -0.705
262 P 0.814
263 E -0.696
264 G 0.196
265 S -0.706
266 H -0.484
267 I -0.044
268 K -0.409
269 D 0.750
270 N -0.422
271 E 0.839
272 Q -0.534
273 A 0.346
274 L 2.176
275 Y -0.389
276 E -0.387
277 L 2.411
278 V -0.210
279 K -0.076
280 C 0.447
281 N 0.551
282 F 1.818
283 D 2.095
284 T -0.084
285 E -0.343
286 E -0.275
287 A 2.643
288 L 0.990
289 R -0.158
290 R -0.623
291 L 0.593
292 R -0.127
293 F -0.940
294 N -0.207
295 V -0.592
296 K -0.462
297 A -0.144
298 A -0.991
299 R -0.815
300 E -0.275
301 E -0.481
302 L -0.515
303 S -0.280
304 V -0.806
305 W 3.648
306 T 1.734
307 E 0.008
308 E 0.415
309 E 2.950
310 C 0.321
311 R -0.130
312 N -0.036
313 F 3.267
314 E 0.138
315 Q -0.113
316 G 2.231
317 L 1.249
318 K -0.741
319 A -0.524
320 Y 1.036
321 G 0.906
322 K 2.526
323 D 1.329
324 F 3.109
325 H -0.553
326 L -0.496
327 I 2.459
328 Q 0.729
329 A 0.348
330 N 0.021
331 K -0.376
332 V 1.883
333 R 0.424
334 T 0.026
335 R 2.277
336 S 0.970
337 V 0.740
338 G 0.474
339 E 0.966
340 C 2.334
341 V 3.235
342 A 0.407
343 F 1.773
344 Y 3.591
345 Y 4.603
346 M -0.277
347 W 1.905
348 K 6.047
349 K 2.401
350 S -0.060
351 E -0.371
352 R 0.213
353 Y -0.044
354 D 0.091
355 F -0.664
356 F -0.282
357 A -0.447
358 Q -0.040
359 Q 0.278
360 T -0.344
361 R 0.092
362 F -0.174
363 G -0.363
364 K -0.084
365 K 0.105
366 K 0.362
367 Y -0.978
368 N -0.613
369 L -0.769
370 H -0.487
371 P 0.103
372 G -0.258
373 V -0.777
374 T -0.370
375 D 0.022
376 Y -0.785
377 M -0.512
378 D -0.116
379 R -0.811
380 L -0.614
381 L -0.440
382 D -0.002
383 E -0.406
384 S -0.697
385 E -0.478
386 S -0.757
387 A -0.708
388 A -0.737
389 S -0.724
390 S -0.591
391 R -0.811
392 A -0.711
393 P -0.389
394 S -0.393
395 P -0.500
396 P -0.774
397 P -0.845
398 T -0.792
399 A -0.932
400 S -0.617
401 N -0.964
402 S -0.715
403 S -0.563
404 N -0.876
405 S -0.682
406 Q -0.616
407 S -0.707
408 E -0.401
409 K -0.530
410 E -0.492
411 D -0.655
412 G -0.783
413 T -0.938
414 V -0.784
415 S -0.432
416 T -0.828
417 A -0.961
418 N -0.552
419 Q -0.548
420 N -0.488
421 G -0.467
422 V -0.515
423 S -0.191
424 S -0.171
425 N -0.282
426 G -0.416
427 P -0.067
428 G -0.460
429 I -0.501
430 L -0.330
431 Q -0.581
432 M -1.000 *
433 L -1.000 *
434 L -1.000 *
435 P -1.000 *
436 V -1.000 *
437 H -1.000 *
438 F -1.000 *
439 S -1.000 *
440 A -1.000 *
441 I -1.000 *
442 S -1.000 *
443 S -1.000 *
444 R -1.000 *
445 A -1.000 *
446 N -1.000 *
447 A -1.000 *
448 F -1.000 *
449 L -1.000 *
450 K -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 228.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MFMFNWFTDC LWTLFLSNYQ PSVESSSPGG SATSDDHEFD PSADMLVHDF DDERTLEEEE
70 80 90 100 110 120
MMEGETNFSS EIEDLAREGD MPIHELLSLY GYGSTVRLPE EDEEEEEEEE EGEDDEDADN
130 140 150 160 170 180
DDNSGCSGEN KEENIKDSSG QEDETQSSND DPSQSVASQD AQEIIRPRRC KYFDTNSEVE
190 200 210 220 230 240
EESEEDEDYI PSEDWKKEIM VGSMFQAEIP VGICRYKENE KVYENDDQLL WDPEYLPEDK
250 260 270 280 290 300
VIIFLKDASR RTGDEKGVEA IPEGSHIKDN EQALYELVKC NFDTEEALRR LRFNVKAARE
310 320 330 340 350 360
ELSVWTEEEC RNFEQGLKAY GKDFHLIQAN KVRTRSVGEC VAFYYMWKKS ERYDFFAQQT
370 380 390 400 410 420
RFGKKKYNLH PGVTDYMDRL LDESESAASS RAPSPPPTAS NSSNSQSEKE DGTVSTANQN
430 440 450
GVSSNGPGIL QMLLPVHFSA ISSRANAFLK
MIER1 is a fibroblast growth factor-activated transcription factor. MIER1 has multiple isoforms as a result of exon skipping, facultative intron usage, etc. Clements et al. showed that one particular isoform with the exon 3A contains a NES at the N-terminus.
[1]. "Differential splicing alters subcellular localization of the alpha but not beta isoform of the MIER1 transcriptional regulator in breast cancer cells"
Clements JA, Mercer FC, Paterno GD, Gillespie LL. (2012)
PLoS One,
7:e32499
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
