Show FASTA Format
>gi|34925430|sp|Q9HCN4.1|GPN1_HUMAN RecName: Full=GPN-loop GTPase 1; AltName: Full=MBD2-interacting protein; Short=MBDin; AltName: Full=XPA-binding protein 1
MAASAAAAELQASGGPRHPVCLLVLGMAGSGKTTFVQRLTGHLHAQGTPPYVINLDPAVHEVPFPANIDI
RDTVKYKEVMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNMSKYVLIDTPGQIEVFTWSASGTIIT
EALASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFIVVMNKTDIIDHSFAVEWMQDFEAFQ
DALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSAVLGTGLDELFVQVTSAAEEYEREYRPEYERLKKSL
ANAESQQQREQLERLRKDMGSVALDAGTAKDSLSPVLHPSDLILTRGTLDEEDEEADSDTDDIDHRVTEE
SHEEPAFQNFMQESMAQYWKRNNK
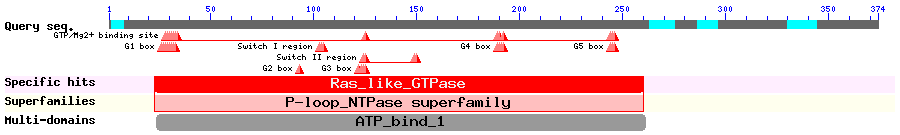
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 987654322348999999628998768998777899999998983499966972776877
Pred: CCCCCCCCCCCCCCCCCCCEEEEEECCCCCCHHHHHHHHHHHHHHCCCCCEEEECCCCCC
AA: MAASAAAAELQASGGPRHPVCLLVLGMAGSGKTTFVQRLTGHLHAQGTPPYVINLDPAVH
10 20 30 40 50 60
Conf: 899886777455336588873519998258999999998607999999997469999399
Pred: CCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCEEE
AA: EVPFPANIDIRDTVKYKEVMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNMSKYVL
70 80 90 100 110 120
Conf: 826992157530334899999986119938999722887568156787799999999751
Pred: EECCCCEEEEECCCCHHHHHHHHHHCCCEEEEEEECCCCCCCHHHHHHHHHHHHHHHHHH
AA: IDTPGQIEVFTWSASGTIITEALASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKT
130 140 150 160 170 180
Conf: 589078730466678234577522779999998013700488999999989985169625
Pred: CCCCEEEEECCCCCCCHHHHHHHHCHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHCCCCE
AA: KLPFIVVMNKTDIIDHSFAVEWMQDFEAFQDALNQETTYVSNLTRSMSLVLDEFYSSLRV
190 200 210 220 230 240
Conf: 883358776899999999999999876413579998988987888899999999996716
Pred: EEEECCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHC
AA: VGVSAVLGTGLDELFVQVTSAAEEYEREYRPEYERLKKSLANAESQQQREQLERLRKDMG
250 260 270 280 290 300
Conf: 885567888898999999970000379999876546899863343467666554358999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHH
AA: SVALDAGTAKDSLSPVLHPSDLILTRGTLDEEDEEADSDTDDIDHRVTEESHEEPAFQNF
310 320 330 340 350 360
Conf: 99999999874089
Pred: HHHHHHHHHHHHCC
AA: MQESMAQYWKRNNK
370
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 A -1.000 *
4 S -1.000 *
5 A -1.000 *
6 A -1.000 *
7 A -1.000 *
8 A -1.000 *
9 E -1.000 *
10 L -1.000 *
11 Q -1.000 *
12 A -1.000 *
13 S -1.000 *
14 G -1.000 *
15 G -1.000 *
16 P -1.000 *
17 R -1.000 *
18 H -0.767
19 P 0.052
20 V -0.651
21 C 0.147
22 L -0.141
23 L 0.479
24 V 0.432
25 L 0.522
26 G 2.743
27 M 1.106
28 A 1.845
29 G 3.089
30 S 1.059
31 G 3.089
32 K 3.089
33 T 2.075
34 T 1.135
35 F 0.926
36 V 0.319
37 Q -0.755
38 R -0.316
39 L 0.531
40 T -0.726
41 G -0.736
42 H -0.567
43 L -0.522
44 H -0.910
45 A -0.905
46 Q -0.915
47 G -0.474
48 T -0.967
49 P -0.899
50 P -0.816
51 Y -0.511
52 V -0.141
53 I 1.088
54 N 2.162
55 L 1.462
56 D 2.442
57 P 2.442
58 A 1.991
59 V 0.209
60 H -1.096
61 E -1.110
62 V -0.532
63 P -0.601
64 F 1.158
65 P -0.817
66 A -0.119
67 N -0.510
68 I 0.201
69 D 1.580
70 I 1.299
71 R -0.039
72 D 0.266
73 T 0.391
74 V 1.225
75 K -0.470
76 Y 0.438
77 K -0.662
78 E -0.364
79 V 0.810
80 M 1.334
81 K -0.682
82 Q -0.585
83 Y -0.461
84 G -0.729
85 L 0.806
86 G 2.345
87 P 2.345
88 N 2.743
89 G 2.032
90 G 1.072
91 I 0.749
92 V 0.673
93 T 0.201
94 S 1.010
95 L 0.572
96 N 0.993
97 L -0.183
98 F 0.860
99 A -0.445
100 T -0.765
101 R 0.126
102 F 0.207
103 D -0.099
104 Q -0.124
105 V 0.091
106 M -0.831
107 K -0.489
108 F -0.900
109 I 0.009
110 E -0.666
111 K -0.750
112 A -0.926
113 Q -1.067
114 N -1.138
115 M -0.998
116 S -1.084
117 K -0.704
118 Y -0.555
119 V 0.004
120 L 1.046
121 I 0.553
122 D 3.089
123 T 0.606
124 P 2.710
125 G 3.089
126 Q 2.711
127 I 1.434
128 E 3.089
129 V 0.383
130 F 1.215
131 T 0.236
132 W 0.136
133 S -0.362
134 A -0.367
135 S 0.067
136 G 0.370
137 T -0.764
138 I -0.567
139 I 0.836
140 T -0.245
141 E -0.692
142 A -0.993
143 L 0.208
144 A -0.727
145 S -0.855
146 S -0.919
147 F -0.455
148 P -0.251
149 T -0.163
150 V -0.165
151 V -0.192
152 I -0.254
153 Y 0.718
154 V 0.546
155 M -0.096
156 D 1.897
157 T 0.143
158 S -0.892
159 R -0.559
160 S -0.102
161 T -0.670
162 N -0.459
163 P -0.167
164 V -0.767
165 T -0.246
166 F 1.496
167 M 0.637
168 S 1.374
169 N 0.143
170 M 0.202
171 L 0.660
172 Y -0.267
173 A 1.055
174 C 0.156
175 S 0.295
176 I 0.035
177 L 1.268
178 Y 0.235
179 K 0.236
180 T -0.277
181 K -0.143
182 L 1.025
183 P 1.196
184 F -0.226
185 I 0.969
186 V -0.038
187 V 0.381
188 M 0.591
189 N 1.267
190 K 2.711
191 T -0.279
192 D 2.495
193 I -0.025
194 I -0.664
195 D -0.571
196 H -1.097
197 S -0.690
198 F -0.917
199 A -1.016
200 V -0.775
201 E -0.700
202 W -0.796
203 M -0.569
204 Q -0.693
205 D 0.330
206 F -0.358
207 E -0.534
208 A -0.972
209 F 0.016
210 Q -0.931
211 D -0.890
212 A -0.982
213 L -0.109
214 N -0.838
215 Q -1.006
216 E -0.927
217 T -0.821
218 T -0.687
219 Y -0.419
220 V -0.476
221 S -0.870
222 N -0.750
223 L 1.022
224 T -0.362
225 R -0.574
226 S -0.022
227 M 0.364
228 S 0.042
229 L -0.681
230 V -0.108
231 L 0.827
232 D 0.052
233 E 0.023
234 F 1.192
235 Y 0.733
236 S -0.776
237 S -0.959
238 L 0.692
239 R -0.853
240 V -0.690
241 V -0.376
242 G -0.575
243 V 0.276
244 S 0.388
245 A -0.556
246 V -0.950
247 L -0.695
248 G -0.033
249 T -0.847
250 G 0.902
251 L -0.423
252 D -0.965
253 E -0.976
254 L -0.171
255 F -0.604
256 V -1.131
257 Q -0.912
258 V 0.749
259 T -0.652
260 S -0.882
261 A -0.419
262 A -0.830
263 E -0.753
264 E -0.333
265 Y -0.127
266 E -1.000
267 R -0.906
268 E -0.869
269 Y -0.457
270 R -0.895
271 P -0.482
272 E -0.697
273 Y -0.617
274 E -0.415
275 R -0.651
276 L -0.935
277 K -0.713
278 K -0.764
279 S -0.727
280 L -0.628
281 A -0.806
282 N -0.860
283 A -0.989
284 E -0.466
285 S -0.841
286 Q -0.989
287 Q -0.867
288 Q -0.803
289 R -0.730
290 E -0.612
291 Q -0.833
292 L -0.557
293 E -0.589
294 R -0.463
295 L -0.679
296 R -0.927
297 K -0.623
298 D -0.227
299 M -0.937
300 G -0.763
301 S -1.103
302 V -0.972
303 A -1.000 *
304 L -1.000 *
305 D -1.000 *
306 A -1.000 *
307 G -1.000 *
308 T -1.000 *
309 A -1.000 *
310 K -1.000 *
311 D -1.000 *
312 S -1.000 *
313 L -1.000 *
314 S -1.000 *
315 P -1.000 *
316 V -1.000 *
317 L -1.000 *
318 H -1.000 *
319 P -1.000 *
320 S -1.000 *
321 D -1.000 *
322 L -1.000 *
323 I -1.000 *
324 L -1.000 *
325 T -1.000 *
326 R -1.000 *
327 G -1.000 *
328 T -1.000 *
329 L -1.000 *
330 D -1.000 *
331 E -1.000 *
332 E -1.000 *
333 D -1.000 *
334 E -1.000 *
335 E -1.000 *
336 A -1.000 *
337 D -1.000 *
338 S -1.000 *
339 D -1.000 *
340 T -1.000 *
341 D -1.000 *
342 D -1.000 *
343 I -1.000 *
344 D -1.000 *
345 H -1.000 *
346 R -1.000 *
347 V -1.000 *
348 T -1.000 *
349 E -1.000 *
350 E -1.000 *
351 S -1.000 *
352 H -1.000 *
353 E -1.000 *
354 E -1.000 *
355 P -1.000 *
356 A -1.000 *
357 F -1.000 *
358 Q -1.000 *
359 N -1.000 *
360 F -1.000 *
361 M -1.000 *
362 Q -1.000 *
363 E -1.000 *
364 S -1.000 *
365 M -1.000 *
366 A -1.000 *
367 Q -1.000 *
368 Y -1.000 *
369 W -1.000 *
370 K -1.000 *
371 R -1.000 *
372 N -1.000 *
373 N -1.000 *
374 K -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 240.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MAASAAAAEL QASGGPRHPV CLLVLGMAGS GKTTFVQRLT GHLHAQGTPP YVINLDPAVH
70 80 90 100 110 120
EVPFPANIDI RDTVKYKEVM KQYGLGPNGG IVTSLNLFAT RFDQVMKFIE KAQNMSKYVL
130 140 150 160 170 180
IDTPGQIEVF TWSASGTIIT EALASSFPTV VIYVMDTSRS TNPVTFMSNM LYACSILYKT
190 200 210 220 230 240
KLPFIVVMNK TDIIDHSFAV EWMQDFEAFQ DALNQETTYV SNLTRSMSLV LDEFYSSLRV
250 260 270 280 290 300
VGVSAVLGTG LDELFVQVTS AAEEYEREYR PEYERLKKSL ANAESQQQRE QLERLRKDMG
310 320 330 340 350 360
SVALDAGTAK DSLSPVLHPS DLILTRGTLD EEDEEADSDT DDIDHRVTEE SHEEPAFQNF
370
MQESMAQYWK RNNK
XAB1/Gpn1 is a GTPase with a glyicine-proline-asparagine structural loop. XAB1/Gpn1 interacts with RNA polymerase II (RNAPII) and is required for the nuclear accumulation of RNAPII. XAB1/Gpn1 is a nucleocytoplasmic shuttling protein. Out of the six putative NESs, only one is surface-exposed as shown by modeling and this is the only one functional.
[1]. "A nuclear export sequence in GPN-loop GTPase 1, an essential protein for nuclear targeting of RNA polymerase II, is necessary and sufficient for nuclear export"
Reyes-Pardo H, Barbosa-Camacho AA, Pérez-Mejía AE, Lara-Chacón B, Salas-Estrada LA, Robledo-Rivera AY, Montero-Morán GM, Lara-González S, Calera MR, Sánchez-Olea R. (2012)
Biochim Biophys Acta,
1823:1756-1766
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
