Nuclear adenylate kinase isoform
UniProt
Show FASTA Format
>tr|Q4DZ57|Q4DZ57_TRYCC Uncharacterized protein OS=Trypanosoma cruzi (strain CL Brener) GN=Tc00.1047053507023.280 PE=4 SV=1
MLQSPKGINILITGTPGTGKTSLAELLAQELGDFKHVEVGKIVKENHFYSEYDNALDTHI
VEEDDEDRLLDFMEPMMVNEGNHVVDYHSSELFPRRWFHLVIVLRASTEVLFERLTARRY
SEQKRDENMEAEIQGLCEEEARGAYDDSIVIVRENNTLEEMAATVDLIRLRVESLRGNAA
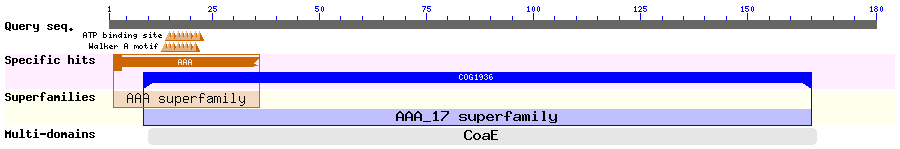
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 999999770899868999878999999988599719974520010698422358557555
Pred: CCCCCCCCEEEEECCCCCCHHHHHHHHHHHHCCCEEEECCCHHHCCCCCCCCCCCCCCCC
AA: MLQSPKGINILITGTPGTGKTSLAELLAQELGDFKHVEVGKIVKENHFYSEYDNALDTHI
10 20 30 40 50 60
Conf: 562246467752201021299836630026888888402899997486679999997249
Pred: CCHHHHHHHHHHHHHHHHCCCCEEEEEECCCCCCCCCCCEEEEEECCCHHHHHHHHHCCC
AA: VEEDDEDRLLDFMEPMMVNEGNHVVDYHSSELFPRRWFHLVIVLRASTEVLFERLTARRY
70 80 90 100 110 120
Conf: 934787779999877579999844899739983699988899999999999998540489
Pred: CHHHHHHHHHHHHHHHHHHHHHHHCCCCEEEECCCCCHHHHHHHHHHHHHHHHHHHHCCC
AA: SEQKRDENMEAEIQGLCEEEARGAYDDSIVIVRENNTLEEMAATVDLIRLRVESLRGNAA
130 140 150 160 170 180
Conf:
Pred:
AA:
Show Conservation Score by AL2CO
1 M -1.000 *
2 L -1.000 *
3 Q -1.000 *
4 S -1.000 *
5 P -1.000 *
6 K -1.000 *
7 G -1.000 *
8 I 0.893
9 N -0.122
10 I 1.483
11 L 0.038
12 I 1.640
13 T 0.728
14 G 3.650
15 T 1.335
16 P 3.439
17 G 2.277
18 T 1.260
19 G 3.230
20 K 4.256
21 T 1.627
22 S -0.223
23 L 0.144
24 A 0.950
25 E -0.550
26 L -0.865
27 L 1.210
28 A -0.613
29 Q -0.302
30 E -0.775
31 L -0.716
32 G -0.943
33 D -0.859
34 F -0.142
35 K -0.674
36 H -0.586
37 V 0.130
38 E -0.460
39 V 0.733
40 G 0.652
41 K -0.312
42 I -0.040
43 V 0.875
44 K -0.420
45 E -0.402
46 N -0.788
47 H -0.355
48 F 0.322
49 Y -0.609
50 S -0.880
51 E -0.701
52 Y -0.407
53 D 0.336
54 N -0.379
55 A -0.761
56 L -0.696
57 D -0.183
58 T 0.406
59 H -0.924
60 I -0.091
61 V 0.668
62 E -1.000 *
63 E -1.000 *
64 D -1.000 *
65 D -0.113
66 E -0.027
67 D 0.002
68 R -0.347
69 L -0.284
70 L -0.435
71 D -0.628
72 F -1.209
73 M -0.344
74 E -0.940
75 P -0.653
76 M -1.127
77 M -0.955
78 V -1.191
79 N -0.845
80 E -0.953
81 G -0.681
82 N -0.864
83 H -1.175
84 V 1.115
85 V 1.417
86 D 0.614
87 Y 0.249
88 H 0.829
89 S -0.750
90 S -0.094
91 E -0.362
92 L -0.678
93 F -0.151
94 P -0.431
95 R -0.714
96 R -0.870
97 W -0.921
98 F 0.304
99 H -0.437
100 L -0.876
101 V 0.645
102 I 0.437
103 V 0.795
104 L 1.272
105 R -0.045
106 A 0.060
107 S -0.353
108 T 0.297
109 E -0.801
110 V -0.874
111 L 1.371
112 F -0.841
113 E -0.545
114 R 1.023
115 L 1.337
116 T -0.711
117 A -0.630
118 R 3.148
119 R 0.104
120 Y 1.337
121 S -0.777
122 E -0.835
123 Q -0.752
124 K 2.439
125 R 0.770
126 D -0.749
127 E 0.035
128 N 1.852
129 M 0.389
130 E 0.107
131 A 1.083
132 E 1.937
133 I -0.123
134 Q 0.326
135 G -0.192
136 L 0.040
137 C 0.811
138 E -0.746
139 E -0.864
140 E 0.911
141 A 1.063
142 R -0.925
143 G -0.167
144 A -1.135
145 Y -0.117
146 D -0.652
147 D -0.880
148 S -0.586
149 I -0.715
150 V 0.413
151 I -0.783
152 V -0.868
153 R 0.243
154 E -0.956
155 N -0.021
156 N -0.549
157 T -0.731
158 L -0.802
159 E -0.610
160 E 0.034
161 M -0.516
162 A -0.704
163 A -0.696
164 T -0.293
165 V 0.055
166 D -0.629
167 L -0.922
168 I 1.194
169 R -0.844
170 L -0.619
171 R -0.268
172 V 0.067
173 E -0.571
174 S -0.594
175 L 0.686
176 R -0.411
177 G -0.769
178 N -0.092
179 A -1.000 *
180 A -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 242.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MLQSPKGINI LITGTPGTGK TSLAELLAQE LGDFKHVEVG KIVKENHFYS EYDNALDTHI
70 80 90 100 110 120
VEEDDEDRLL DFMEPMMVNE GNHVVDYHSS ELFPRRWFHL VIVLRASTEV LFERLTARRY
130 140 150 160 170 180
SEQKRDENME AEIQGLCEEE ARGAYDDSIV IVRENNTLEE MAATVDLIRL RVESLRGNAA
Putative nuclear exportation signal ( with 63% of probability) was identified using the NETNES1.1 predictor.
Beside bioinformatic prediction, no further experiments were performed to validate the putative NES.
TcADKn is shown to be involved in ribosomal 18S RNA processing by direct interaction with ribosomal protein TcRps14.
[1]. "Molecular and Functional Characterization of a Trypanosoma cruzi Nuclear Adenylate Kinase Isoform"
Cámara Mde L, Bouvier LA, Canepa GE, Miranda MR, Pereira CA. (2013)
PLOS Neglected Tropical Diseases,
7(2):
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
