Summary for Neurogenin 3 (NES ID: 243)
Full Name
Neurogenin-3 (NGN-3)
UniProt Alternative Names
Helix-loop-helix protein mATH-4B (mATH4B); Protein atonal homolog 5
Organism
Mus musculus (Mouse)
Experimental Evidence for CRM1-mediated Export
LMB Sensitive, Mislocalization in Xpo1-1 temperature sensitive strains
Mutations That Affect Nuclear Export
*highlighted yellow in the full sequence
L135A
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
*shown as underlined residues in the full sequence
131YIWALTQTLRIA
142
Secondary Structure of Export Signal
Not known
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|3913130|sp|P70661.1|NGN3_MOUSE RecName: Full=Neurogenin-3; Short=NGN-3; AltName: Full=Helix-loop-helix protein mATH-4B; Short=mATH4B; AltName: Full=Protein atonal homolog 5
MAPHPLDALTIQVSPETQQPFPGASDHEVLSSNSTPPSPTLIPRDCSEAEVGDCRGTSRKLRARRGGRNR
PKSELALSKQRRSRRKKANDRERNRMHNLNSALDALRGVLPTFPDDAKLTKIETLRFAHNYIWALTQTLR
IADHSFYGPEPPVPCGELGSPGGGSNGDWGSIYSPVSQAGNLSPTASLEEFPGLQVPSSPSYLLPGALVF
SDFL
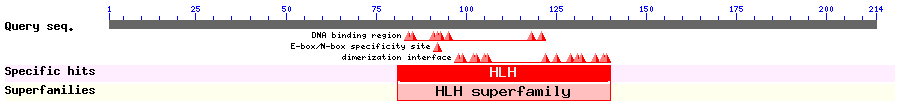
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 999999996568999988999999986556799999999999998641012898774012
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHH
AA: MAPHPLDALTIQVSPETQQPFPGASDHEVLSSNSTPPSPTLIPRDCSEAEVGDCRGTSRK
10 20 30 40 50 60
Conf: 111479999998768888999887756667886422233799998550499999987743
Pred: HHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCH
AA: LRARRGGRNRPKSELALSKQRRSRRKKANDRERNRMHNLNSALDALRGVLPTFPDDAKLT
70 80 90 100 110 120
Conf: 789999999999999998762058899999998988999999999999899888886789
Pred: HHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: KIETLRFAHNYIWALTQTLRIADHSFYGPEPPVPCGELGSPGGGSNGDWGSIYSPVSQAG
130 140 150 160 170 180
Conf: 9999998788999999999998899976567789
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: NLSPTASLEEFPGLQVPSSPSYLLPGALVFSDFL
190 200 210
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 P -1.000 *
4 H -1.000 *
5 P -1.000 *
6 L -1.000 *
7 D -1.000 *
8 A -1.000 *
9 L -1.000 *
10 T -1.000 *
11 I -1.000 *
12 Q -1.000 *
13 V -1.000 *
14 S -1.000 *
15 P -1.000 *
16 E -1.000 *
17 T -1.000 *
18 Q -1.000 *
19 Q -1.000 *
20 P -1.000 *
21 F -1.000 *
22 P -1.000 *
23 G -1.000 *
24 A -1.000 *
25 S -1.000 *
26 D -1.000 *
27 H -1.000 *
28 E -1.000 *
29 V -1.000 *
30 L -0.669
31 S -0.589
32 S -0.589
33 N -0.636
34 S -0.607
35 T -0.624
36 P -0.580
37 P -0.518
38 S -0.600
39 P -0.513
40 T -0.544
41 L -0.673
42 I -0.713
43 P -0.475
44 R -0.598
45 D -0.653
46 C -0.524
47 S -0.626
48 E -0.456
49 A -0.575
50 E -0.440
51 V -0.658
52 G -0.600
53 D -0.626
54 C -0.701
55 R -0.553
56 G -0.528
57 T -0.668
58 S -0.511
59 R -0.510
60 K -0.479
61 L -0.636
62 R -0.575
63 A -0.606
64 R -0.435
65 R -0.606
66 G -0.421
67 G -0.602
68 R -0.523
69 N -0.558
70 R -0.511
71 P -0.510
72 K -0.354
73 S -0.652
74 E -0.519
75 L -0.612
76 A -0.546
77 L -0.627
78 S -0.590
79 K -0.536
80 Q -0.491
81 R -0.399
82 R -0.392
83 S -0.585
84 R 0.342
85 R 2.740
86 K -0.383
87 K -0.160
88 A 1.145
89 N 1.656
90 D -0.152
91 R 2.030
92 E 2.740
93 R 2.740
94 N -0.382
95 R 2.740
96 M 0.948
97 H 0.267
98 N -0.109
99 L 1.193
100 N 2.020
101 S -0.360
102 A 2.358
103 L 1.258
104 D 0.141
105 A -0.245
106 L 2.740
107 R 1.805
108 G -0.425
109 V -0.125
110 L 1.034
111 P 2.356
112 T -0.227
113 F -0.455
114 P 0.051
115 D -0.516
116 D 0.610
117 A 0.087
118 K 1.603
119 L 1.388
120 T 1.988
121 K 2.234
122 I 0.447
123 E 0.651
124 T 1.495
125 L 2.740
126 R 0.731
127 F 0.572
128 A 2.360
129 H -0.205
130 N -0.071
131 Y 2.356
132 I 2.740
133 W -0.493
134 A -0.234
135 L 1.647
136 T -0.452
137 Q -0.224
138 T 0.391
139 L 0.713
140 R -0.333
141 I -0.547
142 A -0.405
143 D -0.183
144 H -0.540
145 S -0.601
146 F -0.640
147 Y -0.566
148 G -0.578
149 P -0.612
150 E -0.612
151 P -0.587
152 P -0.539
153 V -0.686
154 P -0.649
155 C -0.641
156 G -0.611
157 E -0.659
158 L -0.587
159 G -0.559
160 S -0.557
161 P -0.650
162 G -0.591
163 G -0.548
164 G -0.565
165 S -0.597
166 N -0.534
167 G -0.510
168 D -0.513
169 W -0.668
170 G -0.537
171 S -0.376
172 I -1.000 *
173 Y -0.656
174 S -0.336
175 P -0.364
176 V -0.714
177 S -0.480
178 Q -0.560
179 A -0.632
180 G -1.000 *
181 N -1.000 *
182 L -1.000 *
183 S -1.000 *
184 P -1.000 *
185 T -1.000 *
186 A -1.000 *
187 S -1.000 *
188 L -1.000 *
189 E -1.000 *
190 E -1.000 *
191 F -1.000 *
192 P -1.000 *
193 G -1.000 *
194 L -1.000 *
195 Q -1.000 *
196 V -1.000 *
197 P -1.000 *
198 S -1.000 *
199 S -1.000 *
200 P -1.000 *
201 S -1.000 *
202 Y -1.000 *
203 L -1.000 *
204 L -1.000 *
205 P -1.000 *
206 G -1.000 *
207 A -1.000 *
208 L -1.000 *
209 V -1.000 *
210 F -1.000 *
211 S -1.000 *
212 D -1.000 *
213 F -1.000 *
214 L -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 243.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MAPHPLDALT IQVSPETQQP FPGASDHEVL SSNSTPPSPT LIPRDCSEAE VGDCRGTSRK
70 80 90 100 110 120
LRARRGGRNR PKSELALSKQ RRSRRKKAND RERNRMHNLN SALDALRGVL PTFPDDAKLT
130 140 150 160 170 180
KIETLRFAHN YIWALTQTLR IADHSFYGPE PPVPCGELGS PGGGSNGDWG SIYSPVSQAG
190 200 210
NLSPTASLEE FPGLQVPSSP SYLLPGALVF SDFL
3D Structures in PDB
Not Available
Comments
Ngn-3 is a basic-helix-loop-helix (bHLH) protein, which regulates dendritogenesis and synaptogenesis in mouse hippocampal neurons. Ngn-3 is found primarily in the cell nuclear. It is also located in the perikaryon and is found in the cytoplasm in certain stages of neuronal development. Mutation of the NES affected neronal morphology and synaptic inputs.
References
User Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
