RAC-alpha serine/threonine-protein kinase
UniProt
Show FASTA Format
>gi|18027298|gb|AAL55732.1| AKT1 [Homo sapiens]
MSDVAIVKEGWLHKRGEYIKTWRPRYFLLKNDGTFIGYKERPQDVDQREAPLNNFSVAQCQLMKTERPRP
NTFIIRCLQWTTVIERTFHVETPEEREEWTTAIQTVADGLKKQEEEEMDFRSGSPSDNSGAEEMEVSLAK
PKHRVTMNEFEYLKLLGKGTFGKVILVKEKATGRYYAMKILKKEVIVAKDEVAHTLTENRVLQNSRHPFL
TALKYSFQTHDRLCFVMEYANGGELFFHLSRERVFSEDRARFYGAEIVSALDYLHSEKNVVYRDLKLENL
MLDKDGHIKITDFGLCKEGIKDGATMKTFCGTPEYLAPEVLEDNDYGRAVDWWGLGVVMYEMMCGRLPFY
NQDHEKLFELILMEEIRFPRTLGPEAKSLLSGLLKKDPKQRLGGGSEDAKEIMQHRFFAGIVWQHVYEKK
LSPPFKPQVTSETDTRYFDEEFTAQMITITPPDQDDSMECVDSERRPHFPQFSYSASSTA
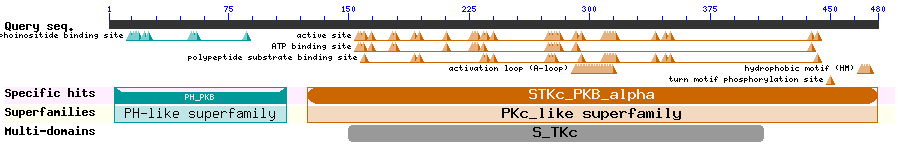
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 987636786336630664333223148994356366553079974578777887310011
Pred: CCCCCEEEEEEEEEECCCCCCCCCCEEEEEECCEEEEEECCCCCCCCCCCCCCCCCCCCC
AA: MSDVAIVKEGWLHKRGEYIKTWRPRYFLLKNDGTFIGYKERPQDVDQREAPLNNFSVAQC
10 20 30 40 50 60
Conf: 100134788984317431003653111137993566778899897630144103555202
Pred: CCCCCCCCCCCCCEEEEEEEEEEEECCCCCCCHHHHHHHHHHHHHHHCCCCCHHHHHHHC
AA: QLMKTERPRPNTFIIRCLQWTTVIERTFHVETPEEREEWTTAIQTVADGLKKQEEEEMDF
70 80 90 100 110 120
Conf: 699999999962000024788876577986237874056743289998827894799983
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCEEEEEECCCCCEEEEEEECCCCCEEEEEE
AA: RSGSPSDNSGAEEMEVSLAKPKHRVTMNEFEYLKLLGKGTFGKVILVKEKATGRYYAMKI
130 140 150 160 170 180
Conf: 201110135513568886877620799824044201126981799997422652000000
Pred: ECCCCCCCCCCHHHHHHHHHHHHCCCCCCEECCCCCCCCCCEEEEEEEEECCCCHHHHHC
AA: LKKEVIVAKDEVAHTLTENRVLQNSRHPFLTALKYSFQTHDRLCFVMEYANGGELFFHLS
190 200 210 220 230 240
Conf: 247678430257999999996301069993530399872000689987860265422799
Pred: CCCCCCCCCHHHHHHHHHHHHHHHHCCCCEEEECCCCCCEECCCCCCEEECCCCCCCCCC
AA: RERVFSEDRARFYGAEIVSALDYLHSEKNVVYRDLKLENLMLDKDGHIKITDFGLCKEGI
250 260 270 280 290 300
Conf: 899842334579654200002569998631014467999988509999987899999999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCHHHHHHH
AA: KDGATMKTFCGTPEYLAPEVLEDNDYGRAVDWWGLGVVMYEMMCGRLPFYNQDHEKLFEL
310 320 330 340 350 360
Conf: 970987789999987999999852369899589999995677509997889957773596
Pred: HHHCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHCCCCCCCCCHHHHHCCC
AA: ILMEEIRFPRTLGPEAKSLLSGLLKKDPKQRLGGGSEDAKEIMQHRFFAGIVWQHVYEKK
370 380 390 400 410 420
Conf: 999988644898877998466542856568888898866578457999999633089999
Pred: CCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCEEECCCCC
AA: LSPPFKPQVTSETDTRYFDEEFTAQMITITPPDQDDSMECVDSERRPHFPQFSYSASSTA
430 440 450 460 470 480
Conf:
Pred:
AA:
Show Conservation Score by AL2CO
1 M -1.000 *
2 S -1.000 *
3 D -1.000 *
4 V -1.000 *
5 A -1.000 *
6 I -1.000 *
7 V -1.000 *
8 K -1.000 *
9 E -1.000 *
10 G -1.000 *
11 W -1.000 *
12 L -1.000 *
13 H -1.000 *
14 K -1.000 *
15 R -1.000 *
16 G -1.000 *
17 E -1.000 *
18 Y -1.000 *
19 I -1.000 *
20 K -1.000 *
21 T -1.000 *
22 W -1.000 *
23 R -1.000 *
24 P -1.000 *
25 R -1.000 *
26 Y -1.000 *
27 F -1.000 *
28 L -1.000 *
29 L -1.000 *
30 K -1.000 *
31 N -1.000 *
32 D -1.000 *
33 G -1.000 *
34 T -1.000 *
35 F -1.000 *
36 I -1.000 *
37 G -1.000 *
38 Y -1.000 *
39 K -1.000 *
40 E -1.000 *
41 R -1.000 *
42 P -1.000 *
43 Q -1.000 *
44 D -1.000 *
45 V -1.000 *
46 D -1.000 *
47 Q -1.000 *
48 R -1.000 *
49 E -1.000 *
50 A -1.000 *
51 P -1.000 *
52 L -1.000 *
53 N -1.000 *
54 N -1.000 *
55 F -1.000 *
56 S -1.000 *
57 V -1.000 *
58 A -1.000 *
59 Q -1.000 *
60 C -1.000 *
61 Q -1.000 *
62 L -1.000 *
63 M -1.000 *
64 K -1.000 *
65 T -1.000 *
66 E -1.000 *
67 R -1.000 *
68 P -1.000 *
69 R -1.000 *
70 P -1.000 *
71 N -1.000 *
72 T -1.000 *
73 F -1.000 *
74 I -1.000 *
75 I -1.000 *
76 R -1.000 *
77 C -1.000 *
78 L -1.000 *
79 Q -1.000 *
80 W -1.000 *
81 T -1.000 *
82 T -1.000 *
83 V -1.000 *
84 I -1.000 *
85 E -1.000 *
86 R -1.000 *
87 T -1.000 *
88 F -1.000 *
89 H -1.000 *
90 V -1.000 *
91 E -1.000 *
92 T -1.000 *
93 P -1.000 *
94 E -1.000 *
95 E -1.000 *
96 R -1.000 *
97 E -1.000 *
98 E -1.000 *
99 W -1.000 *
100 T -0.903
101 T -0.952
102 A -1.059
103 I -1.094
104 Q -0.945
105 T -1.054
106 V -0.986
107 A -1.060
108 D -0.927
109 G -1.065
110 L -1.114
111 K -1.104
112 K -1.072
113 Q -1.074
114 E -0.942
115 E -1.051
116 E -1.081
117 E -1.146
118 M -1.178
119 D -1.189
120 F -1.146
121 R -1.061
122 S -1.178
123 G -1.190
124 S -1.111
125 P -1.003
126 S -0.981
127 D -1.032
128 N -1.030
129 S -1.009
130 G -1.069
131 A -1.115
132 E -0.996
133 E -1.066
134 M -1.078
135 E -0.978
136 V -1.106
137 S -1.066
138 L -1.112
139 A -1.139
140 K -1.043
141 P -0.993
142 K -1.012
143 H -0.958
144 R -0.781
145 V -0.740
146 T -0.704
147 M -0.537
148 N -0.497
149 E -0.312
150 F 1.569
151 E -0.740
152 Y -0.106
153 L -0.418
154 K -0.674
155 L -0.131
156 L 1.169
157 G 1.984
158 K -0.363
159 G 1.774
160 T 0.231
161 F 0.887
162 G 0.911
163 K 0.107
164 V 1.812
165 I -0.318
166 L 0.489
167 V 0.807
168 K -0.304
169 E -0.773
170 K -0.668
171 A -0.826
172 T -0.512
173 G -0.516
174 R -0.880
175 Y -0.555
176 Y 0.386
177 A 2.075
178 M 0.596
179 K 2.450
180 I -0.146
181 L 0.720
182 K -0.337
183 K 1.976
184 E -0.591
185 V -0.870
186 I 0.456
187 V -0.668
188 A -0.593
189 K -0.621
190 D -0.233
191 E -0.338
192 V -0.402
193 A -0.406
194 H 0.059
195 T 0.141
196 L -0.185
197 T -0.626
198 E 1.793
199 N 0.583
200 R -0.783
201 V 0.945
202 L 0.777
203 Q -0.625
204 N -0.919
205 S -0.876
206 R -0.810
207 H -0.064
208 P 1.075
209 F 1.084
210 L 0.817
211 T 0.160
212 A -0.717
213 L 1.027
214 K -0.658
215 Y -0.312
216 S 0.477
217 F 1.310
218 Q 0.877
219 T 0.675
220 H -0.901
221 D -0.625
222 R -0.440
223 L 0.819
224 C 0.762
225 F 0.777
226 V 1.347
227 M 0.532
228 E 0.601
229 Y 0.830
230 A -0.095
231 N -0.628
232 G 2.441
233 G 2.267
234 E 1.104
235 L 1.163
236 F 0.887
237 F -0.706
238 H 0.297
239 L 1.025
240 S -0.605
241 R -0.575
242 E -0.862
243 R -0.526
244 V -0.824
245 F 1.060
246 S -0.703
247 E -0.303
248 D -0.849
249 R -0.593
250 A 0.652
251 R -0.231
252 F -0.057
253 Y 0.909
254 G -0.204
255 A 0.106
256 E 0.047
257 I 0.923
258 V -0.115
259 S -0.383
260 A 0.881
261 L 0.688
262 D -0.921
263 Y 0.251
264 L 1.035
265 H 2.388
266 S -0.646
267 E -0.697
268 K -0.658
269 N 0.048
270 V 0.882
271 V 0.710
272 Y 1.357
273 R 2.441
274 D 2.442
275 L 1.378
276 K 2.078
277 L 1.591
278 E 1.271
279 N 2.442
280 L 1.055
281 M 1.212
282 L 0.551
283 D -0.105
284 K -1.045
285 D -0.778
286 G 0.440
287 H 0.248
288 I 0.773
289 K -0.196
290 I 1.130
291 T 0.544
292 D 2.442
293 F 1.405
294 G 2.441
295 L 0.932
296 C 1.119
297 K 2.234
298 E -0.739
299 G -0.848
300 I -0.156
301 K -1.159
302 D -0.922
303 G -0.571
304 A -0.742
305 T -0.858
306 M 0.289
307 K -0.939
308 T 1.696
309 F 0.473
310 C 1.168
311 G 1.791
312 T 1.432
313 P 0.291
314 E 0.408
315 Y 1.472
316 L 0.848
317 A 1.789
318 P 2.441
319 E 2.089
320 V 0.837
321 L 1.207
322 E -0.745
323 D -0.390
324 N -0.858
325 D -0.434
326 Y 1.723
327 G 0.505
328 R -0.891
329 A -0.721
330 V 1.167
331 D 2.483
332 W 1.187
333 W 2.323
334 G 0.746
335 L 0.441
336 G 2.862
337 V 0.497
338 V 0.946
339 M 0.084
340 Y 1.578
341 E 0.879
342 M 1.646
343 M 0.360
344 C -0.432
345 G 1.295
346 R -0.980
347 L 0.163
348 P 2.479
349 F 1.547
350 Y -0.971
351 N -0.253
352 Q -0.646
353 D 0.332
354 H -0.760
355 E -0.639
356 K -0.619
357 L 0.520
358 F 0.001
359 E -0.534
360 L -0.603
361 I 1.617
362 L -0.539
363 M -0.981
364 E -0.387
365 E -0.644
366 I 0.215
367 R -1.081
368 F 0.129
369 P -0.103
370 R -0.884
371 T -1.031
372 L -0.022
373 G -0.194
374 P -0.964
375 E -0.786
376 A 0.154
377 K -0.768
378 S -0.586
379 L 0.242
380 L 0.795
381 S -0.716
382 G -0.459
383 L 1.235
384 L 1.235
385 K -0.695
386 K 0.429
387 D -0.395
388 P -0.318
389 K -0.989
390 Q -0.796
391 R 2.862
392 L 0.963
393 G 1.292
394 G -0.416
395 G -0.761
396 S -0.854
397 E -0.941
398 D -0.222
399 A -0.912
400 K -0.941
401 E -0.481
402 I 0.731
403 M -0.204
404 Q -1.005
405 H 0.586
406 R -0.839
407 F 1.135
408 F 0.941
409 A -0.727
410 G -0.964
411 I 0.190
412 V -0.325
413 W 1.075
414 Q -0.896
415 H -0.682
416 V 0.250
417 Y -1.051
418 E -0.803
419 K 0.093
420 K -0.251
421 L -0.047
422 S -0.705
423 P 0.653
424 P 0.135
425 F 0.520
426 K -0.339
427 P 1.555
428 Q -1.013
429 V -0.212
430 T -0.912
431 S -0.263
432 E -0.810
433 T -0.945
434 D 0.131
435 T -0.432
436 R -0.785
437 Y 0.101
438 F 1.414
439 D 0.483
440 E -0.648
441 E -0.778
442 F 0.642
443 T 0.678
444 A -0.941
445 Q -0.542
446 M -0.840
447 I -0.382
448 T -0.935
449 I -0.785
450 T -0.272
451 P -0.794
452 P -1.017
453 D -0.849
454 Q -0.934
455 D -1.034
456 D -1.076
457 S -0.955
458 M -1.087
459 E -0.923
460 C -1.038
461 V -0.569
462 D -0.864
463 S -0.943
464 E -1.017
465 R -0.993
466 R -0.714
467 P -0.972
468 H -1.020
469 F 1.165
470 P -0.938
471 Q 0.222
472 F 1.713
473 S 0.705
474 Y 0.894
475 S -0.516
476 A -0.384
477 S -0.202
478 S -1.000 *
479 T -1.000 *
480 A -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - ../alignments/261.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MSDVAIVKEG WLHKRGEYIK TWRPRYFLLK NDGTFIGYKE RPQDVDQREA PLNNFSVAQC
70 80 90 100 110 120
QLMKTERPRP NTFIIRCLQW TTVIERTFHV ETPEEREEWT TAIQTVADGL KKQEEEEMDF
130 140 150 160 170 180
RSGSPSDNSG AEEMEVSLAK PKHRVTMNEF EYLKLLGKGT FGKVILVKEK ATGRYYAMKI
190 200 210 220 230 240
LKKEVIVAKD EVAHTLTENR VLQNSRHPFL TALKYSFQTH DRLCFVMEYA NGGELFFHLS
250 260 270 280 290 300
RERVFSEDRA RFYGAEIVSA LDYLHSEKNV VYRDLKLENL MLDKDGHIKI TDFGLCKEGI
310 320 330 340 350 360
KDGATMKTFC GTPEYLAPEV LEDNDYGRAV DWWGLGVVMY EMMCGRLPFY NQDHEKLFEL
370 380 390 400 410 420
ILMEEIRFPR TLGPEAKSLL SGLLKKDPKQ RLGGGSEDAK EIMQHRFFAG IVWQHVYEKK
430 440 450 460 470 480
LSPPFKPQVT SETDTRYFDE EFTAQMITIT PPDQDDSMEC VDSERRPHFP QFSYSASSTA
3QKL (X-Ray,1.90 Å resolution)
4EJN (X-Ray,2.19 Å resolution)
Atk is a central signaling molecule in the PI3 Kinase pathway and is associated with cancer cell invasion. Saji et al. reported that nuclear Akt1 initiates distinct downstream events sufficient to induce cell migration. They identified a conserved nuclear export signal in the kinase domain (AA 272-284), which may contribute to the regulation of Akt1 localization. The mechanism of Akt1 nuclear import is still unknown.
[1]. "Akt1 contains a functional leucine-rich nuclear export sequence"
Saji M1, Vasko V, Kada F, Allbritton EH, Burman KD, Ringel MD. (2005)
Biochem Biophys Res Commun,
332:167-73
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
