Receptor-interacting serine/threonine-protein kinase 3
UniProt
Show FASTA Format
>gi|205371831|sp|Q9Y572.2|RIPK3_HUMAN RecName: Full=Receptor-interacting serine/threonine-protein kinase 3; AltName: Full=RIP-like protein kinase 3; AltName: Full=Receptor-interacting protein 3; Short=RIP-3
MSCVKLWPSGAPAPLVSIEELENQELVGKGGFGTVFRAQHRKWGYDVAVKIVNSKAISREVKAMASLDNE
FVLRLEGVIEKVNWDQDPKPALVTKFMENGSLSGLLQSQCPRPWPLLCRLLKEVVLGMFYLHDQNPVLLH
RDLKPSNVLLDPELHVKLADFGLSTFQGGSQSGTGSGEPGGTLGYLAPELFVNVNRKASTASDVYSFGIL
MWAVLAGREVELPTEPSLVYEAVCNRQNRPSLAELPQAGPETPGLEGLKELMQLCWSSEPKDRPSFQECL
PKTDEVFQMVENNMNAAVSTVKDFLSQLRSSNRRFSIPESGQGGTEMDGFRRTIENQHSRNDVMVSEWLN
KLNLEEPPSSVPKKCPSLTKRSRAQEEQVPQAWTAGTSSDSMAQPPQTPETSTFRNQMPSPTSTGTPSPG
PRGNQGAERQGMNWSCRTPEPNPVTGRPLVNIYNCSGVQVGDNNYLTMQQTTALPTWGLAPSGKGRGLQH
PPPVGSQEGPKDPEAWSRPQGWYNHSGK
Show Domain Info by CDD
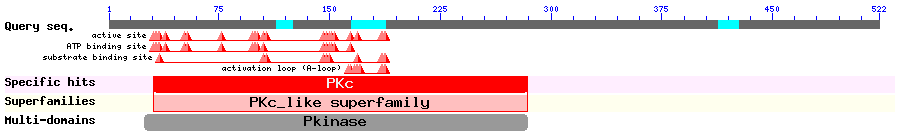
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 975568999999996271102786144377776378644356793699981282648999
Pred: CCCCCCCCCCCCCCCCCHHHCCCCEEECCCCCCEEEEEEECCCCCEEEEEECCHHHHHHH
AA: MSCVKLWPSGAPAPLVSIEELENQELVGKGGFGTVFRAQHRKWGYDVAVKIVNSKAISRE
10 20 30 40 50 60
Conf: 999960599952668322430477899996388423678896543014999986688879
Pred: HHHHHHCCCCCEEEEEEEEECCCCCCCCCCEEEEECCCCCCHHHHCCCCCCCCHHHHHHH
AA: VKAMASLDNEFVLRLEGVIEKVNWDQDPKPALVTKFMENGSLSGLLQSQCPRPWPLLCRL
70 80 90 100 110 120
Conf: 999996522001699971214799998212899956426665321227876668888998
Pred: HHHHHHHHHHCCCCCCCEEECCCCCCCCCCCCCCCEEECCCCCCCCCCCCCCCCCCCCCC
AA: LKEVVLGMFYLHDQNPVLLHRDLKPSNVLLDPELHVKLADFGLSTFQGGSQSGTGSGEPG
130 140 150 160 170 180
Conf: 553223763333368899877863115799999972897899999550688841089999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCCEEEEEECCCCCC
AA: GTLGYLAPELFVNVNRKASTASDVYSFGILMWAVLAGREVELPTEPSLVYEAVCNRQNRP
190 200 210 220 230 240
Conf: 999899999999886999999998327899999990233487999999998768876532
Pred: CCCCCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHH
AA: SLAELPQAGPETPGLEGLKELMQLCWSSEPKDRPSFQECLPKTDEVFQMVENNMNAAVST
250 260 270 280 290 300
Conf: 221100012466657878899999877888788223267886204666311999999999
Pred: HHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHCCCCCCCCCC
AA: VKDFLSQLRSSNRRFSIPESGQGGTEMDGFRRTIENQHSRNDVMVSEWLNKLNLEEPPSS
310 320 330 340 350 360
Conf: 998899960443103246887788645899989999999885577899999999999999
Pred: CCCCCCCCHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: VPKKCPSLTKRSRAQEEQVPQAWTAGTSSDSMAQPPQTPETSTFRNQMPSPTSTGTPSPG
370 380 390 400 410 420
Conf: 999999878899888999999999999914742673100047853565477889888999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCEEEECCCCCEECCCCEEEECCCCCCCCCCCC
AA: PRGNQGAERQGMNWSCRTPEPNPVTGRPLVNIYNCSGVQVGDNNYLTMQQTTALPTWGLA
430 440 450 460 470 480
Conf: 99999999999999999999998887799876665899
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: PSGKGRGLQHPPPVGSQEGPKDPEAWSRPQGWYNHSGK
490 500 510
Show Conservation Score by AL2CO
1 M -1.000 *
2 S -1.000 *
3 C -1.000 *
4 V -1.000 *
5 K -1.000 *
6 L -1.000 *
7 W -1.000 *
8 P -1.000 *
9 S -1.000 *
10 G -1.000 *
11 A -1.000 *
12 P -1.000 *
13 A -1.000 *
14 P -1.000 *
15 L -1.000 *
16 V -0.847
17 S 0.229
18 I -0.700
19 E -0.443
20 E -0.819
21 L 1.049
22 E -0.902
23 N -1.127
24 Q -1.250
25 E -0.709
26 L -0.397
27 V 0.691
28 G 1.794
29 K -0.540
30 G 2.222
31 G 1.060
32 F 1.168
33 G 2.222
34 T -0.881
35 V 1.775
36 F 0.976
37 R 0.073
38 A 1.149
39 Q -0.992
40 H 0.306
41 R -1.125
42 K -0.645
43 W -1.000 *
44 G -0.066
45 Y -0.965
46 D -0.682
47 V 0.783
48 A 1.891
49 V 0.957
50 K 2.222
51 I -0.197
52 V 0.401
53 N -0.255
54 S -0.906
55 K -0.709
56 A -0.115
57 I 0.470
58 S -0.553
59 R -0.176
60 E 2.222
61 V 0.095
62 K -0.966
63 A -0.386
64 M 0.193
65 A -0.552
66 S -0.289
67 L 0.952
68 D 0.032
69 N 0.703
70 E -0.587
71 F 0.880
72 V 0.929
73 L 0.750
74 R -0.470
75 L 0.559
76 E -0.328
77 G 1.304
78 V 0.040
79 I 0.184
80 E -0.760
81 K -1.000 *
82 V -1.000 *
83 N -1.000 *
84 W -1.000 *
85 D -1.000 *
86 Q -0.478
87 D -0.888
88 P -1.099
89 K 0.092
90 P -0.631
91 A 0.025
92 L 1.169
93 V 0.383
94 T 0.583
95 K 0.052
96 F 1.063
97 M 0.718
98 E -0.491
99 N 0.770
100 G -0.291
101 S 1.396
102 L 1.839
103 S -0.142
104 G -1.204
105 L -0.429
106 L 0.660
107 Q -0.246
108 S -0.547
109 Q -0.983
110 C -1.148
111 P -0.967
112 R 0.027
113 P -0.807
114 W 1.396
115 P -1.144
116 L -0.770
117 L -0.096
118 C -0.789
119 R -0.940
120 L 0.730
121 L 0.163
122 K -1.229
123 E 0.498
124 V 0.451
125 V 0.246
126 L 0.003
127 G 1.842
128 M 0.504
129 F -0.547
130 Y 1.793
131 L 1.328
132 H 2.222
133 D -0.078
134 Q -0.410
135 N -0.626
136 P -0.221
137 V -0.589
138 L 0.953
139 L 0.965
140 H 1.913
141 R 0.949
142 D 1.913
143 L 0.622
144 K 2.222
145 P 0.137
146 S 0.556
147 N 2.222
148 V 1.211
149 L 2.222
150 L 1.307
151 D 1.022
152 P -1.179
153 E -0.499
154 L 0.180
155 H -0.559
156 V 0.467
157 K 1.071
158 L 0.820
159 A 0.633
160 D 2.222
161 F 1.914
162 G 1.914
163 L 0.280
164 S 1.280
165 T 0.645
166 F -0.535
167 Q -0.844
168 G -1.154
169 G -1.052
170 S -0.493
171 Q -0.788
172 S -1.104
173 G -0.876
174 T -0.725
175 G -1.072
176 S -0.173
177 G -0.997
178 E -0.464
179 P -0.308
180 G -0.152
181 G 1.078
182 T 1.589
183 L -0.136
184 G 0.202
185 Y 1.491
186 L 0.670
187 A 0.734
188 P 1.891
189 E 2.222
190 L 0.209
191 F -1.000 *
192 V -1.000 *
193 N -0.286
194 V -0.771
195 N -0.789
196 R 0.065
197 K -1.014
198 A -0.028
199 S 0.617
200 T -0.857
201 A 0.111
202 S 0.901
203 D 1.891
204 V 0.447
205 Y 1.388
206 S 1.076
207 F 1.148
208 G 1.539
209 I 0.946
210 L 0.275
211 M 0.004
212 W 0.581
213 A 0.713
214 V 0.466
215 L 0.382
216 A 0.219
217 G 0.582
218 R -0.366
219 E -0.207
220 V -0.692
221 E -1.157
222 L -1.327
223 P -1.260
224 T -1.246
225 E -1.110
226 P -1.230
227 S -1.152
228 L -1.095
229 V -0.886
230 Y -1.180
231 E -1.185
232 A -1.037
233 V -0.848
234 C -1.142
235 N -1.145
236 R -1.125
237 Q -1.019
238 N -1.101
239 R -0.920
240 P -1.044
241 S -1.080
242 L -0.984
243 A -1.137
244 E -0.898
245 L -0.838
246 P -0.798
247 Q -1.120
248 A -1.170
249 G -1.121
250 P -1.196
251 E -1.198
252 T -1.313
253 P -1.041
254 G -1.169
255 L -1.111
256 E -1.185
257 G -0.898
258 L -0.397
259 K -0.634
260 E -0.752
261 L 0.318
262 M 0.113
263 Q -0.408
264 L -0.772
265 C 1.757
266 W -0.004
267 S -0.190
268 S -0.888
269 E -1.084
270 P -0.117
271 K -1.183
272 D -1.006
273 R 2.222
274 P 1.641
275 S -0.837
276 F 0.109
277 Q -0.942
278 E -0.888
279 C 0.123
280 L -0.758
281 P -1.058
282 K -0.808
283 T 0.386
284 D -0.536
285 E -1.000 *
286 V -1.000 *
287 F -1.000 *
288 Q -1.000 *
289 M -1.000 *
290 V -1.000 *
291 E -1.000 *
292 N -1.000 *
293 N -1.000 *
294 M -1.000 *
295 N -1.000 *
296 A -1.000 *
297 A -1.000 *
298 V -1.000 *
299 S -1.000 *
300 T -1.000 *
301 V -1.000 *
302 K -1.000 *
303 D -1.000 *
304 F -1.000 *
305 L -1.000 *
306 S -1.000 *
307 Q -1.000 *
308 L -1.000 *
309 R -1.000 *
310 S -1.000 *
311 S -1.000 *
312 N -1.000 *
313 R -1.000 *
314 R -1.000 *
315 F -1.000 *
316 S -1.000 *
317 I -1.000 *
318 P -1.000 *
319 E -1.000 *
320 S -1.000 *
321 G -1.000 *
322 Q -1.000 *
323 G -1.000 *
324 G -1.000 *
325 T -1.000 *
326 E -1.000 *
327 M -1.000 *
328 D -1.000 *
329 G -1.000 *
330 F -1.000 *
331 R -1.000 *
332 R -1.000 *
333 T -1.000 *
334 I -1.000 *
335 E -1.000 *
336 N -1.000 *
337 Q -1.000 *
338 H -1.000 *
339 S -1.000 *
340 R -1.000 *
341 N -1.000 *
342 D -1.000 *
343 V -1.000 *
344 M -1.000 *
345 V -1.000 *
346 S -1.000 *
347 E -1.000 *
348 W -1.000 *
349 L -1.000 *
350 N -1.000 *
351 K -1.000 *
352 L -1.000 *
353 N -1.000 *
354 L -1.000 *
355 E -1.000 *
356 E -1.000 *
357 P -1.000 *
358 P -1.000 *
359 S -1.000 *
360 S -1.000 *
361 V -1.000 *
362 P -1.000 *
363 K -1.000 *
364 K -1.000 *
365 C -1.000 *
366 P -1.000 *
367 S -1.000 *
368 L -1.000 *
369 T -1.000 *
370 K -1.000 *
371 R -1.000 *
372 S -1.000 *
373 R -1.000 *
374 A -1.000 *
375 Q -1.000 *
376 E -1.000 *
377 E -1.000 *
378 Q -1.000 *
379 V -1.000 *
380 P -1.000 *
381 Q -1.000 *
382 A -1.000 *
383 W -1.000 *
384 T -1.000 *
385 A -1.000 *
386 G -1.000 *
387 T -1.000 *
388 S -1.000 *
389 S -1.000 *
390 D -1.000 *
391 S -1.000 *
392 M -1.000 *
393 A -1.000 *
394 Q -1.000 *
395 P -1.000 *
396 P -1.000 *
397 Q -1.000 *
398 T -1.000 *
399 P -1.000 *
400 E -1.000 *
401 T -1.000 *
402 S -1.000 *
403 T -1.000 *
404 F -1.000 *
405 R -1.000 *
406 N -1.000 *
407 Q -1.000 *
408 M -1.000 *
409 P -1.000 *
410 S -1.000 *
411 P -1.000 *
412 T -1.000 *
413 S -1.000 *
414 T -1.000 *
415 G -1.000 *
416 T -1.000 *
417 P -1.000 *
418 S -1.000 *
419 P -1.000 *
420 G -1.000 *
421 P -1.000 *
422 R -1.000 *
423 G -1.000 *
424 N -1.000 *
425 Q -1.000 *
426 G -1.000 *
427 A -1.000 *
428 E -1.000 *
429 R -1.000 *
430 Q -1.000 *
431 G -1.000 *
432 M -1.000 *
433 N -1.000 *
434 W -1.000 *
435 S -1.000 *
436 C -1.000 *
437 R -1.000 *
438 T -1.000 *
439 P -1.000 *
440 E -1.000 *
441 P -1.000 *
442 N -1.000 *
443 P -1.000 *
444 V -1.000 *
445 T -1.000 *
446 G -1.000 *
447 R -1.000 *
448 P -1.000 *
449 L -1.000 *
450 V -1.000 *
451 N -1.000 *
452 I -1.000 *
453 Y -1.000 *
454 N -1.000 *
455 C -1.000 *
456 S -1.000 *
457 G -1.000 *
458 V -1.000 *
459 Q -1.000 *
460 V -1.000 *
461 G -1.000 *
462 D -1.000 *
463 N -1.000 *
464 N -1.000 *
465 Y -1.000 *
466 L -1.000 *
467 T -1.000 *
468 M -1.000 *
469 Q -1.000 *
470 Q -1.000 *
471 T -1.000 *
472 T -1.000 *
473 A -1.000 *
474 L -1.000 *
475 P -1.000 *
476 T -1.000 *
477 W -1.000 *
478 G -1.000 *
479 L -1.000 *
480 A -1.000 *
481 P -1.000 *
482 S -1.000 *
483 G -1.000 *
484 K -1.000 *
485 G -1.000 *
486 R -1.000 *
487 G -1.000 *
488 L -1.000 *
489 Q -1.000 *
490 H -1.000 *
491 P -1.000 *
492 P -1.000 *
493 P -1.000 *
494 V -1.000 *
495 G -1.000 *
496 S -1.000 *
497 Q -1.000 *
498 E -1.000 *
499 G -1.000 *
500 P -1.000 *
501 K -1.000 *
502 D -1.000 *
503 P -1.000 *
504 E -1.000 *
505 A -1.000 *
506 W -1.000 *
507 S -1.000 *
508 R -1.000 *
509 P -1.000 *
510 Q -1.000 *
511 G -1.000 *
512 W -1.000 *
513 Y -1.000 *
514 N -1.000 *
515 H -1.000 *
516 S -1.000 *
517 G -1.000 *
518 K -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 32.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MSCVKLWPSG APAPLVSIEE LENQELVGKG GFGTVFRAQH RKWGYDVAVK IVNSKAISRE
70 80 90 100 110 120
VKAMASLDNE FVLRLEGVIE KVNWDQDPKP ALVTKFMENG SLSGLLQSQC PRPWPLLCRL
130 140 150 160 170 180
LKEVVLGMFY LHDQNPVLLH RDLKPSNVLL DPELHVKLAD FGLSTFQGGS QSGTGSGEPG
190 200 210 220 230 240
GTLGYLAPEL FVNVNRKAST ASDVYSFGIL MWAVLAGREV ELPTEPSLVY EAVCNRQNRP
250 260 270 280 290 300
SLAELPQAGP ETPGLEGLKE LMQLCWSSEP KDRPSFQECL PKTDEVFQMV ENNMNAAVST
310 320 330 340 350 360
VKDFLSQLRS SNRRFSIPES GQGGTEMDGF RRTIENQHSR NDVMVSEWLN KLNLEEPPSS
370 380 390 400 410 420
VPKKCPSLTK RSRAQEEQVP QAWTAGTSSD SMAQPPQTPE TSTFRNQMPS PTSTGTPSPG
430 440 450 460 470 480
PRGNQGAERQ GMNWSCRTPE PNPVTGRPLV NIYNCSGVQV GDNNYLTMQQ TTALPTWGLA
490 500 510
PSGKGRGLQH PPPVGSQEGP KDPEAWSRPQ GWYNHSGK
RIP3 belongs to the receptor-interacting protein Ser/Thr kinase family and has been characterized as a pro-aptotic protein involved in the tumor necrosis factor receptor-1 signaling complex. RIP3 shuttles with an unconventional NLS and two NESs. In addition, it has a NES at the N-terminus independent of CRM1 pathway (104-142).
[1]. "Nucleocytoplasmic shuttling of receptor-interacting protein 3 (RIP3): identification of novel nuclear export and import signals in RIP3."
Yang Y, Ma J, Chen Y, Wu M. (2004)
J Biol Chem,
279:38820-9
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
