Summary for RNF146 (NES ID: 322)
Full Name
E3 ubiquitin-protein ligase RNF146
UniProt Alternative Names
Dactylidin, Iduna, RING finger protein 146, RING-type E3 ubiquitin transferase RNF146
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Unknown
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
Undetermined
Secondary Structure of Export Signal
Unknown
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>sp|Q9NTX7|RN146_HUMAN E3 ubiquitin-protein ligase RNF146 OS=Homo sapiens OX=9606 GN=RNF146 PE=1 SV=1
MMAGCGEIDHSINMLPTNRKANESCSNTAPSLTVPECAICLQTCVHPVSLPCKHVFCYLC
VKGASWLGKRCALCRQEIPEDFLDKPTLLSPEELKAASRGNGEYAWYYEGRNGWWQYDER
TSRELEDAFSKGKKNTEMLIAGFLYVADLENMVQYRRNEHGRRRKIKRDIIDIPKKGVAG
LRLDCDANTVNLARESSADGADSVSAQSGASVQPLVSSVRPLTSVDGQLTSPATPSPDAS
TSLEDSFAHLQLSGDNTAERSHRGEGEEDHESPSSGRVPAPDTSIEETESDASSDSEDVS
AVVAQHSLTQQRLLVSNANQTVPDRSDRSGTDRSVAGGGTVSVSVRSRRPDGQCTVTEV
Show Domain Info by CDD
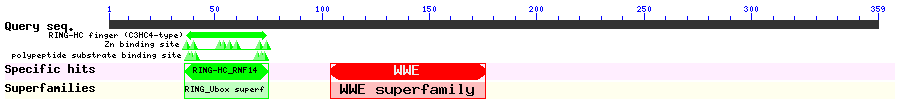
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 987555633553555114754110016789779887843347677876469998121345
Pred: CCCCCCCCCCCCCCCCCHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHH
AA: MMAGCGEIDHSINMLPTNRKANESCSNTAPSLTVPECAICLQTCVHPVSLPCKHVFCYLC
10 20 30 40 50 60
Conf: 655434288776422235743458984357467764204887404423026874123577
Pred: HHHHHHCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHCCCCCCCCCCCC
AA: VKGASWLGKRCALCRQEIPEDFLDKPTLLSPEELKAASRGNGEYAWYYEGRNGWWQYDER
70 80 90 100 110 120
Conf: 337898740699971476351010332010000246799873224323578978017860
Pred: CHHHHHHHHCCCCCCHHHHHHCCEEEECCCCCCCCCCCCCCCCCCCCCCCCCCCEEEEEC
AA: TSRELEDAFSKGKKNTEMLIAGFLYVADLENMVQYRRNEHGRRRKIKRDIIDIPKKGVAG
130 140 150 160 170 180
Conf: 014457776675436898998875668998756546889999866888899999999987
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: LRLDCDANTVNLARESSADGADSVSAQSGASVQPLVSSVRPLTSVDGQLTSPATPSPDAS
190 200 210 220 230 240
Conf: 714545765213899964458899887667899999888999996433467888852024
Pred: CCHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHH
AA: TSLEDSFAHLQLSGDNTAERSHRGEGEEDHESPSSGRVPAPDTSIEETESDASSDSEDVS
250 260 270 280 290 300
Conf: 65510103455431038887899999999999989999974334446799987202439
Pred: HHHHCCHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCEEEC
AA: AVVAQHSLTQQRLLVSNANQTVPDRSDRSGTDRSVAGGGTVSVSVRSRRPDGQCTVTEV
310 320 330 340 350
Show Conservation Score by AL2CO
1 M -1.000 *
2 M -1.000 *
3 A -1.000 *
4 G -1.000 *
5 C -1.000 *
6 G -1.000 *
7 E -1.000 *
8 I -1.000 *
9 D 1.181
10 H -1.000 *
11 S 1.015
12 I -0.404
13 N 0.130
14 M -0.421
15 L -0.529
16 P 0.062
17 T -0.286
18 N -0.464
19 R -0.317
20 K -0.769
21 A -1.082
22 N -0.450
23 E -0.483
24 S -0.881
25 C -1.093
26 S -0.613
27 N -0.815
28 T -1.187
29 A -0.539
30 P -0.593
31 S -0.570
32 L -0.848
33 T -0.605
34 V -0.381
35 P -0.234
36 E 0.149
37 C 3.501
38 A 0.764
39 I 2.017
40 C 3.501
41 L 1.590
42 Q 0.683
43 T -0.621
44 C 0.009
45 V -0.383
46 H 0.393
47 P 2.995
48 V 0.644
49 S -0.259
50 L 1.529
51 P 2.307
52 C 2.974
53 K -0.747
54 H 2.974
55 V 0.751
56 F 2.707
57 C 2.974
58 Y 0.195
59 L 0.790
60 C 2.518
61 V 0.859
62 K 1.070
63 G 0.909
64 A -0.199
65 S 0.505
66 W -0.738
67 L -0.326
68 G 0.462
69 K 0.568
70 R 0.857
71 C 3.501
72 A 2.020
73 L 1.255
74 C 3.501
75 R 2.334
76 Q -0.346
77 E -0.649
78 I 1.284
79 P 0.859
80 E -0.862
81 D -0.205
82 F 0.751
83 L 0.322
84 D -0.129
85 K -0.129
86 P 1.873
87 T -0.712
88 L 1.633
89 L 0.839
90 S -0.765
91 P -0.872
92 E -0.782
93 E 0.068
94 L -0.833
95 K -0.635
96 A -1.058
97 A -1.143
98 S -0.856
99 R -0.874
100 G -0.577
101 N -0.740
102 G -0.712
103 E -0.596
104 Y -0.229
105 A -0.515
106 W 1.565
107 Y 1.451
108 Y 1.548
109 E 0.743
110 G 1.080
111 R 0.034
112 N 0.559
113 G 0.990
114 W 2.153
115 W 1.548
116 Q 0.183
117 Y 1.333
118 D 1.726
119 E -0.609
120 R 0.609
121 T 0.799
122 S 0.391
123 R -1.185
124 E 0.681
125 L 0.544
126 E 1.947
127 D -0.698
128 A -0.326
129 F 0.261
130 S -0.593
131 K -0.620
132 G -0.371
133 K -0.361
134 K -0.380
135 N -0.824
136 T -0.282
137 E -0.091
138 M 0.420
139 L 0.885
140 I 1.373
141 A 1.031
142 G 1.210
143 F -0.004
144 L 0.221
145 Y 1.959
146 V -0.103
147 A 0.269
148 D 1.364
149 L 0.813
150 E -0.415
151 N -0.548
152 M 0.071
153 V -0.678
154 Q 0.837
155 Y -0.667
156 R 1.178
157 R 0.308
158 N -0.081
159 E -0.220
160 H -0.045
161 G -0.299
162 R 1.141
163 R -0.338
164 R 1.685
165 K 0.227
166 I 0.550
167 K 0.630
168 R 1.271
169 D 1.175
170 I -0.835
171 I -1.180
172 D -0.009
173 I -0.390
174 P -0.012
175 K -0.134
176 K 0.925
177 G 1.114
178 V 1.406
179 A 0.592
180 G 1.797
181 L 1.285
182 R 0.824
183 L -0.072
184 D -0.598
185 C -1.272
186 D -0.563
187 A -1.079
188 N -0.815
189 T -0.934
190 V -1.099
191 N -0.722
192 L -0.751
193 A -0.767
194 R -0.452
195 E -1.001
196 S -0.526
197 S -0.883
198 A -0.931
199 D -0.565
200 G -0.673
201 A -0.905
202 D -0.627
203 S -0.438
204 V -0.940
205 S -0.826
206 A -0.806
207 Q -0.969
208 S -0.856
209 G -0.675
210 A -0.524
211 S -0.605
212 V -1.043
213 Q -0.928
214 P -0.608
215 L -0.904
216 V -0.841
217 S -0.381
218 S -0.442
219 V -0.739
220 R -0.534
221 P -0.475
222 L -0.538
223 T -0.293
224 S -0.574
225 V -0.655
226 D -0.874
227 G -0.877
228 Q -0.687
229 L -0.837
230 T -0.647
231 S -0.582
232 P -0.802
233 A -0.517
234 T -0.866
235 P -0.304
236 S -0.925
237 P -0.469
238 D -0.489
239 A -0.581
240 S -0.734
241 T -0.792
242 S -0.075
243 L 0.225
244 E -0.671
245 D -0.037
246 S -0.194
247 F 0.251
248 A -0.468
249 H 0.196
250 L -0.043
251 Q -0.393
252 L 0.049
253 S -0.241
254 G -0.989
255 D -0.449
256 N -0.590
257 T -0.886
258 A -0.760
259 E -0.710
260 R -0.735
261 S -0.767
262 H -0.669
263 R -0.650
264 G -0.670
265 E -0.670
266 G -0.781
267 E -0.587
268 E -0.283
269 D -0.668
270 H -0.546
271 E -0.639
272 S -0.242
273 P -0.720
274 S -0.311
275 S -0.736
276 G -0.559
277 R -0.776
278 V -0.987
279 P -0.590
280 A -0.610
281 P -0.774
282 D -0.513
283 T -0.760
284 S -0.802
285 I -0.895
286 E -0.328
287 E -0.364
288 T -0.644
289 E -0.379
290 S -0.557
291 D -0.464
292 A -1.000 *
293 S -0.366
294 S -0.117
295 D 0.577
296 S -1.000 *
297 E -1.000 *
298 D -1.000 *
299 V -1.000 *
300 S -1.000 *
301 A -1.000 *
302 V -1.000 *
303 V -1.000 *
304 A -1.000 *
305 Q -1.000 *
306 H -1.000 *
307 S -1.000 *
308 L -1.000 *
309 T -1.000 *
310 Q -1.000 *
311 Q -1.000 *
312 R -1.000 *
313 L -1.000 *
314 L -1.000 *
315 V -1.000 *
316 S -1.000 *
317 N -1.000 *
318 A -1.000 *
319 N -1.000 *
320 Q -1.000 *
321 T -1.000 *
322 V -1.000 *
323 P -1.000 *
324 D -1.000 *
325 R -1.000 *
326 S -1.000 *
327 D -1.000 *
328 R -1.000 *
329 S -1.000 *
330 G -1.000 *
331 T -1.000 *
332 D -1.000 *
333 R -1.000 *
334 S -1.000 *
335 V -1.000 *
336 A -1.000 *
337 G -1.000 *
338 G -1.000 *
339 G -1.000 *
340 T -1.000 *
341 V -1.000 *
342 S -1.000 *
343 V -1.000 *
344 S -1.000 *
345 V -1.000 *
346 R -1.000 *
347 S -1.000 *
348 R -1.000 *
349 R -1.000 *
350 P -1.000 *
351 D -1.000 *
352 G -1.000 *
353 Q -1.000 *
354 C -1.000 *
355 T -1.000 *
356 V -1.000 *
357 T -1.000 *
358 E -1.000 *
359 V -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 322.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MMAGCGEIDH SINMLPTNRK ANESCSNTAP SLTVPECAIC LQTCVHPVSL PCKHVFCYLC
70 80 90 100 110 120
VKGASWLGKR CALCRQEIPE DFLDKPTLLS PEELKAASRG NGEYAWYYEG RNGWWQYDER
130 140 150 160 170 180
TSRELEDAFS KGKKNTEMLI AGFLYVADLE NMVQYRRNEH GRRRKIKRDI IDIPKKGVAG
190 200 210 220 230 240
LRLDCDANTV NLARESSADG ADSVSAQSGA SVQPLVSSVR PLTSVDGQLT SPATPSPDAS
250 260 270 280 290 300
TSLEDSFAHL QLSGDNTAER SHRGEGEEDH ESPSSGRVPA PDTSIEETES DASSDSEDVS
310 320 330 340 350
AVVAQHSLTQ QRLLVSNANQ TVPDRSDRSG TDRSVAGGGT VSVSVRSRRP DGQCTVTEV
3D Structures in PDB
2D8T (NMR)
3V3L (X-Ray,1.6 Å resolution)
6CF6 (X-Ray,1.9 Å resolution)
Comments
RNF146 is an E3 ubiquitin-protein ligase that mediates the ubiquitination of poly-ADP-ribosylated (PARsylated) proteins involved in cell survival and various stress response. Sheng et al. identified a putative NES in 243-252, which matches classical NES consensus and is conserved in human, mouse, dog, xenopus, and zebrafish. CRM1 dependence was demonstrated by KPT-185 inhibition (instead of LMB) and co-IP with CRM1.
References
[1]. "XPO1-mediated Nuclear Export of RNF146 Protects From Angiotensin II-induced Endothelial Cellular Injury"
Sheng Z, Xu Y, Wang S, Yuan Y, Huang T, Lu P. (2018)
Biochem Biophys Res Commun,
503(3):1544-1549
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
