Serine/threonine-protein kinase UL13
UniProt
Show FASTA Format
>sp|P04290|UL13_HHV11 Serine/threonine-protein kinase UL13 OS=Human herpesvirus 1 (strain 17) OX=10299 GN=UL13 PE=1 SV=1
MDESRRQRPAGHVAANLSPQGARQRSFKDWLASYVHSNPHGASGRPSGPSLQDAAVSRSS
HGSRHRSGLRERLRAGLSRWRMSRSSHRRASPETPGTAAKLNRPPLRRSQAALTAPPSSP
SHILTLTRIRKLCSPVFAINPALHYTTLEIPGARSFGGSGGYGDVQLIREHKLAVKTIKE
KEWFAVELIATLLVGECVLRAGRTHNIRGFIAPLGFSLQQRQIVFPAYDMDLGKYIGQLA
SLRTTNPSVSTALHQCFTELARAVVFLNTTCGISHLDIKCANILVMLRSDAVSLRRAVLA
DFSLVTLNSNSTIARGQFCLQEPDLKSPRMFGMPTALTTANFHTLVGHGYNQPPELLVKY
LNNERAEFTNHRLKHDVGLAVDLYALGQTLLELVVSVYVAPSLGVPVTRFPGYQYFNNQL
SPDFALALLAYRCVLHPALFVNSAETNTHGLAYDVPEGIRRHLRNPKIRRAFTDRCINYQ
HTHKAILSSVALPPELKPLLVLVSRLCHTNPCARHALS
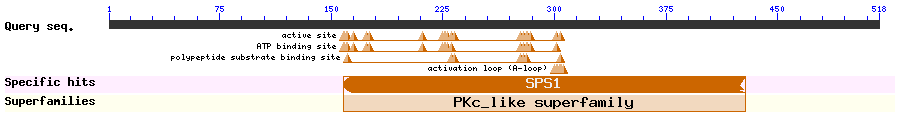
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 998899999944345779998888874111210136999999999999999888888999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: MDESRRQRPAGHVAANLSPQGARQRSFKDWLASYVHSNPHGASGRPSGPSLQDAAVSRSS
10 20 30 40 50 60
Conf: 999999984000025888778898899999999999966689998775789888999999
Pred: CCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: HGSRHRSGLRERLRAGLSRWRMSRSSHRRASPETPGTAAKLNRPPLRRSQAALTAPPSSP
70 80 90 100 110 120
Conf: 522023200235797544799962110347986655444556289993498289999604
Pred: CCCHHHHHHCCCCCCCCCCCCCCCEEECCCCCCCCCCCCCCCCEEEEECCCEEEEEEEEC
AA: SHILTLTRIRKLCSPVFAINPALHYTTLEIPGARSFGGSGGYGDVQLIREHKLAVKTIKE
130 140 150 160 170 180
Conf: 663088999999998899873687775860426331003613564234489245541026
Pred: CCCHHHHHHHHHHHHHHHHHHCCCCCCCCEEEEEEEECCCCEEEEECCCCCHHHHHHCCC
AA: KEWFAVELIATLLVGECVLRAGRTHNIRGFIAPLGFSLQQRQIVFPAYDMDLGKYIGQLA
190 200 210 220 230 240
Conf: 569999630789999999999999985205981313799995786158994234452760
Pred: CCCCCCCCHHHHHHHHHHHHHHHHHHHHHCCCEEECCCCCCCEEECCCCCCCCCCCEEEC
AA: SLRTTNPSVSTALHQCFTELARAVVFLNTTCGISHLDIKCANILVMLRSDAVSLRRAVLA
250 260 270 280 290 300
Conf: 123402078996433228962899899863377776556763103402589997899987
Pred: CCCCCEECCCCCCCCCEEEECCCCCCCCCCCCCCCCCCCCCCCCEEECCCCCCHHHHHHH
AA: DFSLVTLNSNSTIARGQFCLQEPDLKSPRMFGMPTALTTANFHTLVGHGYNQPPELLVKY
310 320 330 340 350 360
Conf: 608655334678664025678799988899999998413589996434578743345788
Pred: HHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC
AA: LNNERAEFTNHRLKHDVGLAVDLYALGQTLLELVVSVYVAPSLGVPVTRFPGYQYFNNQL
370 380 390 400 410 420
Conf: 857798886766845101115896543358787671001002589889999999999811
Pred: CHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCHHCCCCCHHHHHHHHHHHHHHH
AA: SPDFALALLAYRCVLHPALFVNSAETNTHGLAYDVPEGIRRHLRNPKIRRAFTDRCINYQ
430 440 450 460 470 480
Conf: 00122100146998616699999852068933525589
Pred: HHHHHHHCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCC
AA: HTHKAILSSVALPPELKPLLVLVSRLCHTNPCARHALS
490 500 510
Show Conservation Score by AL2CO
1 M -1.000 *
2 D -1.000 *
3 E -1.000 *
4 S -1.000 *
5 R -1.000 *
6 R -1.000 *
7 Q -1.000 *
8 R -1.000 *
9 P -1.000 *
10 A -1.000 *
11 G -1.000 *
12 H -1.000 *
13 V -1.000 *
14 A -1.000 *
15 A -1.000 *
16 N -1.000 *
17 L -1.000 *
18 S -1.000 *
19 P -1.000 *
20 Q -1.000 *
21 G -1.000 *
22 A -1.000 *
23 R -1.000 *
24 Q -1.000 *
25 R -0.041
26 S -1.000 *
27 F -1.000 *
28 K -1.000 *
29 D -1.000 *
30 W -1.000 *
31 L -1.000 *
32 A -1.000 *
33 S -1.000 *
34 Y -1.000 *
35 V -1.000 *
36 H -0.227
37 S -0.297
38 N -0.673
39 P -0.359
40 H -0.538
41 G -0.264
42 A -0.301
43 S -0.603
44 G -0.711
45 R -0.543
46 P -0.529
47 S -0.572
48 G -0.541
49 P -0.666
50 S -0.316
51 L -0.487
52 Q -0.647
53 D -0.565
54 A -0.847
55 A -0.546
56 V -0.534
57 S -0.793
58 R -0.110
59 S -0.340
60 S -0.331
61 H -0.347
62 G -0.479
63 S -0.780
64 R -0.537
65 H -0.830
66 R -0.768
67 S -0.134
68 G -0.386
69 L -0.443
70 R -0.572
71 E -0.422
72 R -0.477
73 L -0.523
74 R -0.563
75 A -0.625
76 G 0.074
77 L -0.918
78 S -0.419
79 R -0.689
80 W -0.896
81 R -0.079
82 M -0.952
83 S -0.460
84 R -0.515
85 S -0.618
86 S -0.446
87 H -0.551
88 R -0.737
89 R -0.762
90 A -0.624
91 S -0.300
92 P -0.587
93 E -0.590
94 T -0.572
95 P -0.529
96 G -0.714
97 T -0.607
98 A -0.568
99 A -0.797
100 K -0.924
101 L -0.643
102 N -0.690
103 R -0.424
104 P -0.428
105 P -0.440
106 L -0.679
107 R -0.830
108 R -0.555
109 S -0.312
110 Q -0.624
111 A -0.380
112 A -0.533
113 L -0.462
114 T -0.813
115 A -0.882
116 P -0.811
117 P 0.073
118 S -0.663
119 S -0.768
120 P -0.504
121 S -0.637
122 H -0.746
123 I -0.105
124 L -0.620
125 T -0.805
126 L -0.650
127 T -0.816
128 R -0.686
129 I -0.510
130 R -0.540
131 K -0.644
132 L -0.557
133 C -0.753
134 S -0.808
135 P -0.557
136 V -0.885
137 F -0.694
138 A -0.721
139 I -0.353
140 N -0.412
141 P -0.050
142 A -0.703
143 L -0.674
144 H -0.875
145 Y -0.563
146 T -0.505
147 T -0.656
148 L -0.713
149 E -0.513
150 I -0.412
151 P -0.536
152 G -0.250
153 A -0.715
154 R -0.401
155 S -0.524
156 F -0.233
157 G 1.071
158 G 2.828
159 S 0.163
160 G 3.759
161 G 1.052
162 Y 2.140
163 G 2.525
164 D -0.377
165 V 3.250
166 Q -0.256
167 L -0.305
168 I -0.496
169 R -0.198
170 E -0.221
171 H -0.421
172 K -0.785
173 L 0.737
174 A 2.394
175 V 1.142
176 K 3.754
177 T -0.078
178 I 0.220
179 K -0.495
180 E -0.339
181 K -0.994
182 E -0.389
183 W -0.273
184 F -0.493
185 A -0.735
186 V -0.880
187 E 0.694
188 L -0.057
189 I -0.085
190 A -0.248
191 T -0.205
192 L -0.341
193 L 0.529
194 V 0.102
195 G -0.177
196 E -0.184
197 C -0.380
198 V -0.351
199 L 0.035
200 R -0.083
201 A -0.330
202 G -0.576
203 R -0.598
204 T -0.571
205 H -0.768
206 N -0.441
207 I -0.393
208 R -0.787
209 G 0.011
210 F 1.238
211 I 0.582
212 A -0.824
213 P -0.024
214 L 0.148
215 G 0.006
216 F -0.201
217 S 0.403
218 L -0.283
219 Q -0.593
220 Q -0.548
221 R -0.333
222 Q -0.597
223 I 0.345
224 V 0.285
225 F 0.626
226 P -0.062
227 A -0.036
228 Y 0.232
229 D -0.680
230 M -0.160
231 D 0.802
232 L 0.393
233 G -0.451
234 K -0.435
235 Y 1.121
236 I -0.631
237 G -0.530
238 Q 0.054
239 L 0.429
240 A -0.524
241 S -0.833
242 L -0.956
243 R -0.502
244 T -0.712
245 T -0.802
246 N -0.802
247 P -0.665
248 S -0.803
249 V -0.907
250 S -0.389
251 T -0.700
252 A -0.799
253 L -0.536
254 H -0.542
255 Q -0.238
256 C -0.644
257 F 0.808
258 T -0.690
259 E 0.552
260 L 0.902
261 A -0.186
262 R 0.155
263 A 2.704
264 V 1.809
265 V -0.705
266 F 3.016
267 L 2.627
268 N 3.018
269 T -0.692
270 T -0.527
271 C 0.190
272 G 0.688
273 I 1.919
274 S 0.268
275 H 3.756
276 L 0.754
277 D 3.753
278 I 1.940
279 K 1.936
280 C 0.752
281 A 0.035
282 N 4.320
283 I 2.391
284 L 2.751
285 V 1.982
286 M 0.023
287 L -0.619
288 R -0.469
289 S -0.638
290 D -0.339
291 A -0.420
292 V -0.699
293 S -0.953
294 L 0.429
295 R -0.086
296 R -0.042
297 A 0.753
298 V 0.923
299 L 2.147
300 A 1.177
301 D 3.721
302 F 1.631
303 S 2.701
304 L 0.705
305 V 0.938
306 T -0.083
307 L 0.356
308 N 0.064
309 S -0.002
310 N 0.141
311 S 1.333
312 T -1.000 *
313 I -1.000 *
314 A -1.000 *
315 R 0.001
316 G -0.269
317 Q -0.350
318 F -0.030
319 C -0.664
320 L 0.142
321 Q -0.803
322 E -0.582
323 P -0.073
324 D -0.412
325 L -0.549
326 K -0.533
327 S -0.259
328 P -0.492
329 R -0.583
330 M -0.715
331 F -0.558
332 G -0.301
333 M -0.346
334 P -0.659
335 T -0.659
336 A -1.000 *
337 L -1.000 *
338 T -1.000 *
339 T -1.000 *
340 A -1.000 *
341 N -1.000 *
342 F -1.000 *
343 H -1.000 *
344 T -1.000 *
345 L 0.551
346 V 0.672
347 G -1.000 *
348 H -1.000 *
349 G -1.000 *
350 Y -0.211
351 N -0.007
352 Q 0.779
353 P 2.103
354 P 0.566
355 E 0.237
356 L 0.897
357 L 0.793
358 V -0.294
359 K -0.193
360 Y 0.006
361 L -1.000 *
362 N -1.000 *
363 N -1.000 *
364 E -1.000 *
365 R -1.000 *
366 A -1.000 *
367 E -1.000 *
368 F -1.000 *
369 T -1.000 *
370 N -1.000 *
371 H -1.000 *
372 R -1.000 *
373 L -1.000 *
374 K -1.000 *
375 H -0.214
376 D 0.190
377 V -0.169
378 G 1.340
379 L 0.345
380 A 0.036
381 V 0.910
382 D 3.695
383 L 1.158
384 Y 0.774
385 A 1.365
386 L 1.393
387 G 0.933
388 Q 0.377
389 T 0.511
390 L 1.410
391 L 0.349
392 E 1.138
393 L 0.549
394 V -1.000 *
395 V -1.000 *
396 S -1.000 *
397 V -1.000 *
398 Y -1.000 *
399 V -1.000 *
400 A -1.000 *
401 P -1.000 *
402 S -1.000 *
403 L -1.000 *
404 G -1.000 *
405 V -1.000 *
406 P -1.000 *
407 V -1.000 *
408 T -1.000 *
409 R -1.000 *
410 F -1.000 *
411 P -1.000 *
412 G -1.000 *
413 Y -1.000 *
414 Q -1.000 *
415 Y -1.000 *
416 F -1.000 *
417 N -1.000 *
418 N -1.000 *
419 Q -1.000 *
420 L -1.000 *
421 S -1.000 *
422 P -1.000 *
423 D -1.000 *
424 F -1.000 *
425 A -1.000 *
426 L -1.000 *
427 A -1.000 *
428 L -1.000 *
429 L -1.000 *
430 A -1.000 *
431 Y -1.000 *
432 R -1.000 *
433 C -1.000 *
434 V -1.000 *
435 L -1.000 *
436 H -1.000 *
437 P -1.000 *
438 A -1.000 *
439 L -1.000 *
440 F -1.000 *
441 V -1.000 *
442 N -1.000 *
443 S -1.000 *
444 A -1.000 *
445 E -1.000 *
446 T -1.000 *
447 N -1.000 *
448 T -1.000 *
449 H -1.000 *
450 G -1.000 *
451 L -1.000 *
452 A -1.000 *
453 Y -1.000 *
454 D -1.000 *
455 V -1.000 *
456 P -1.000 *
457 E -1.000 *
458 G -1.000 *
459 I -1.000 *
460 R -1.000 *
461 R -1.000 *
462 H -1.000 *
463 L -1.000 *
464 R -1.000 *
465 N -1.000 *
466 P -1.000 *
467 K -1.000 *
468 I -1.000 *
469 R -1.000 *
470 R -1.000 *
471 A -1.000 *
472 F -1.000 *
473 T -1.000 *
474 D -1.000 *
475 R -1.000 *
476 C -1.000 *
477 I -1.000 *
478 N -1.000 *
479 Y -1.000 *
480 Q -1.000 *
481 H -1.000 *
482 T -1.000 *
483 H -1.000 *
484 K -1.000 *
485 A -1.000 *
486 I -1.000 *
487 L -1.000 *
488 S -1.000 *
489 S -1.000 *
490 V -1.000 *
491 A -1.000 *
492 L -1.000 *
493 P -1.000 *
494 P -1.000 *
495 E -1.000 *
496 L -1.000 *
497 K -1.000 *
498 P -1.000 *
499 L -1.000 *
500 L -1.000 *
501 V -1.000 *
502 L -1.000 *
503 V -1.000 *
504 S -1.000 *
505 R -1.000 *
506 L -1.000 *
507 C -1.000 *
508 H -1.000 *
509 T -1.000 *
510 N -1.000 *
511 P -1.000 *
512 C -1.000 *
513 A -1.000 *
514 R -1.000 *
515 H -1.000 *
516 A -1.000 *
517 L -1.000 *
518 S -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 330.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MDESRRQRPA GHVAANLSPQ GARQRSFKDW LASYVHSNPH GASGRPSGPS LQDAAVSRSS
70 80 90 100 110 120
HGSRHRSGLR ERLRAGLSRW RMSRSSHRRA SPETPGTAAK LNRPPLRRSQ AALTAPPSSP
130 140 150 160 170 180
SHILTLTRIR KLCSPVFAIN PALHYTTLEI PGARSFGGSG GYGDVQLIRE HKLAVKTIKE
190 200 210 220 230 240
KEWFAVELIA TLLVGECVLR AGRTHNIRGF IAPLGFSLQQ RQIVFPAYDM DLGKYIGQLA
250 260 270 280 290 300
SLRTTNPSVS TALHQCFTEL ARAVVFLNTT CGISHLDIKC ANILVMLRSD AVSLRRAVLA
310 320 330 340 350 360
DFSLVTLNSN STIARGQFCL QEPDLKSPRM FGMPTALTTA NFHTLVGHGY NQPPELLVKY
370 380 390 400 410 420
LNNERAEFTN HRLKHDVGLA VDLYALGQTL LELVVSVYVA PSLGVPVTRF PGYQYFNNQL
430 440 450 460 470 480
SPDFALALLA YRCVLHPALF VNSAETNTHG LAYDVPEGIR RHLRNPKIRR AFTDRCINYQ
490 500 510
HTHKAILSSV ALPPELKPLL VLVSRLCHTN PCARHALS
pUL13 is a multifunctional serine/threonine kinase that has diverse role in viral infection. Funk et al. found this protein to have nuclear export activity using Nuclear EXport Trapped by RAPamycin (NEX-TRAP). EBV homolog pBGLF4 was shown to have both nuclear and cytoplasmic localization, but did not show any nuclear export activity in NEX-TRAP assay.
[1]. "Comprehensive analysis of nuclear export of herpes simplex virus type 1 tegument proteins and their Epstein-Barr virus orthologs"
Funk C, Raschbichler V, Lieber D, Wetschky J, Arnold EK, Leimser J, Biggel M, Friedel CC, Ruzsics Z, Bailer SM. (2019)
Traffic,
20:152-167
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
