Summary for HECTD1 (NES ID: 334)
Full Name
E3 ubiquitin-protein ligase HECTD1 UniProt
Alternative Names
E3 ligase for inhibin receptor, EULIR, HECT domain-containing protein 1, HECT-type E3 ubiquitin transferase HECTD1
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Unknown
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
Undetermined
Secondary Structure of Export Signal
Unknown
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>sp|Q9ULT8|HECD1_HUMAN E3 ubiquitin-protein ligase HECTD1 OS=Homo sapiens OX=9606 GN=HECTD1 PE=1 SV=3 MADVDPDTLLEWLQMGQGDERDMQLIALEQLCMLLLMSDNVDRCFETCPPRTFLPALCKI FLDESAPDNVLEVTARAITYYLDVSAECTRRIVGVDGAIKALCNRLVVVELNNRTSRDLA EQCVKVLELICTRESGAVFEAGGLNCVLTFIRDSGHLVHKDTLHSAMAVVSRLCGKMEPQ DSSLEICVESLSSLLKHEDHQVSDGALRCFASLADRFTRRGVDPAPLAKHGLTEELLSRM AAAGGTVSGPSSACKPGRSTTGAPSTTADSKLSNQVSTIVSLLSTLCRGSPVVTHDLLRS ELPDSIESALQGDERCVLDTMRLVDLLLVLLFEGRKALPKSSAGSTGRIPGLRRLDSSGE RSHRQLIDCIRSKDTDALIDAIDTGAFEVNFMDDVGQTLLNWASAFGTQEMVEFLCERGA DVNRGQRSSSLHYAACFGRPQVAKTLLRHGANPDLRDEDGKTPLDKARERGHSEVVAILQ SPGDWMCPVNKGDDKKKKDTNKDEEECNEPKGDPEMAPIYLKRLLPVFAQTFQQTMLPSI RKASLALIRKMIHFCSEALLKEVCDSDVGHNLPTILVEITATVLDQEDDDDGHLLALQII RDLVDKGGDIFLDQLARLGVISKVSTLAGPSSDDENEEESKPEKEDEPQEDAKELQQGKP YHWRDWSIIRGRDCLYIWSDAAALELSNGSNGWFRFILDGKLATMYSSGSPEGGSDSSES RSEFLEKLQRARGQVKPSTSSQPILSAPGPTKLTVGNWSLTCLKEGEIAIHNSDGQQATI LKEDLPGFVFESNRGTKHSFTAETSLGSEFVTGWTGKRGRKLKSKLEKTKQKVRTMARDL YDDHFKAVESMPRGVVVTLRNIATQLESSWELHTNRQCIESENTWRDLMKTALENLIVLL KDENTISPYEMCSSGLVQALLTVLNNSMDLDMKQDCSQLVERINVFKTAFSENEDDESRP AVALIRKLIAVLESIERLPLHLYDTPGSTYNLQILTRRLRFRLERAPGETALIDRTGRML KMEPLATVESLEQYLLKMVAKQWYDFDRSSFVFVRKLREGQNFIFRHQHDFDENGIIYWI GTNAKTAYEWVNPAAYGLVVVTSSEGRNLPYGRLEDILSRDNSALNCHSNDDKNAWFAID LGLWVIPSAYTLRHARGYGRSALRNWVFQVSKDGQNWTSLYTHVDDCSLNEPGSTATWPL DPPKDEKQGWRHVRIKQMGKNASGQTHYLSLSGFELYGTVNGVCEDQLGKAAKEAEANLR RQRRLVRSQVLKYMVPGARVIRGLDWKWRDQDGSPQGEGTVTGELHNGWIDVTWDAGGSN SYRMGAEGKFDLKLAPGYDPDTVASPKPVSSTVSGTTQSWSSLVKNNCPDKTSAAAGSSS RKGSSSSVCSVASSSDISLGSTKTERRSEIVMEHSIVSGADVHEPIVVLSSAENVPQTEV GSSSSASTSTLTAETGSENAERKLGPDSSVRTPGESSAISMGIVSVSSPDVSSVSELTNK EAASQRPLSSSASNRLSVSSLLAAGAPMSSSASVPNLSSRETSSLESFVRRVANIARTNA TNNMNLSRSSSDNNTNTLGRNVMSTATSPLMGAQSFPNLTTPGTTSTVTMSTSSVTSSSN VATATTVLSVGQSLSNTLTTSLTSTSSESDTGQEAEYSLYDFLDSCRASTLLAELDDDED LPEPDEEDDENEDDNQEDQEYEEVMILRRPSLQRRAGSRSDVTHHAVTSQLPQVPAGAGS RPIGEQEEEEYETKGGRRRTWDDDYVLKRQFSALVPAFDPRPGRTNVQQTTDLEIPPPGT PHSELLEEVECTPSPRLALTLKVTGLGTTREVELPLTNFRSTIFYYVQKLLQLSCNGNVK SDKLRRIWEPTYTIMYREMKDSDKEKENGKMGCWSIEHVEQYLGTDELPKNDLITYLQKN ADAAFLRHWKLTGTNKSIRKNRNCSQLIAAYKDFCEHGTKSGLNQGAISTLQSSDILNLT KEQPQAKAGNGQNSCGVEDVLQLLRILYIVASDPYSRISQEDGDEQLQFTFPPDEFTSKK ITTKILQQIEEPLALASGALPDWCEQLTSKCPFLIPFETRQLYFTCTAFGASRAIVWLQN RREATVERTRTTSSVRRDDPGEFRVGRLKHERVKVPRGESLMEWAENVMQIHADRKSVLE VEFLGEEGTGLGPTLEFYALVAAEFQRTDLGAWLCDDNFPDDESRHVDLGGGLKPPGYYV QRSCGLFTAPFPQDSDELERITKLFHFLGIFLAKCIQDNRLVDLPISKPFFKLMCMGDIK SNMSKLIYESRGDRDLHCTESQSEASTEEGHDSLSVGSFEEDSKSEFILDPPKPKPPAWF NGILTWEDFELVNPHRARFLKEIKDLAIKRRQILSNKGLSEDEKNTKLQELVLKNPSGSG PPLSIEDLGLNFQFCPSSRIYGFTAVDLKPSGEDEMITMDNAEEYVDLMFDFCMHTGIQK QMEAFRDGFNKVFPMEKLSSFSHEEVQMILCGNQSPSWAAEDIINYTEPKLGYTRDSPGF LRFVRVLCGMSSDERKAFLQFTTGCSTLPPGGLANLHPRLTVVRKVDATDASYPSVNTCV HYLKLPEYSSEEIMRERLLAATMEKGFHLNShow Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 998995679999852888613569999999999986147511001338997655677787
Pred: CCCCCHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHH
AA: MADVDPDTLLEWLQMGQGDERDMQLIALEQLCMLLLMSDNVDRCFETCPPRTFLPALCKI
10 20 30 40 50 60
Conf: 303899930765522211223231743333221103259999622112147762317789
Pred: HCCCCCCCHHHHHHHHHEEEEEECCCCCCCCEECCCCHHHHHHHHHHHCCCCCCCCHHHH
AA: FLDESAPDNVLEVTARAITYYLDVSAECTRRIVGVDGAIKALCNRLVVVELNNRTSRDLA
70 80 90 100 110 120
Conf: 989899886520466420314885236799804896303533433559999861577889
Pred: HHHHHHHHHHHCCCCCCHHCCCCCCEEEEEEECCCCCCHHHHHHHHHHHHHHHHCCCCCC
AA: EQCVKVLELICTRESGAVFEAGGLNCVLTFIRDSGHLVHKDTLHSAMAVVSRLCGKMEPQ
130 140 150 160 170 180
Conf: 983679999987421157975540355544454655441399950222003199999999
Pred: CCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHCCCCCHHHHHHCHHHHHHHHH
AA: DSSLEICVESLSSLLKHEDHQVSDGALRCFASLADRFTRRGVDPAPLAKHGLTEELLSRM
190 200 210 220 230 240
Conf: 832899889998877999889999878875577754689999864206994666898617
Pred: HHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHCCCCHHHHHHHHHC
AA: AAAGGTVSGPSSACKPGRSTTGAPSTTADSKLSNQVSTIVSLLSTLCRGSPVVTHDLLRS
250 260 270 280 290 300
Conf: 605899998328964010012388999997622788777777898898887656799534
Pred: CHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: ELPDSIESALQGDERCVLDTMRLVDLLLVLLFEGRKALPKSSAGSTGRIPGLRRLDSSGE
310 320 330 340 350 360
Conf: 320256787632793899999870975333456420458866641092999999982296
Pred: CCHHHHHHHHHCCCHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHCCCHHHHHHHHHCCC
AA: RSHRQLIDCIRSKDTDALIDAIDTGAFEVNFMDDVGQTLLNWASAFGTQEMVEFLCERGA
370 380 390 400 410 420
Conf: 445565565101455307458899998259999997768999204655107288888741
Pred: CCCCCCCCCCHHHHHHCCCHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHCCHHHHHHHHC
AA: DVNRGQRSSSLHYAACFGRPQVAKTLLRHGANPDLRDEDGKTPLDKARERGHSEVVAILQ
430 440 450 460 470 480
Conf: 988754568888854557778886545799899863899999665799998421027427
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHCCCCHH
AA: SPGDWMCPVNKGDDKKKKDTNKDEEECNEPKGDPEMAPIYLKRLLPVFAQTFQQTMLPSI
490 500 510 520 530 540
Conf: 898999899998607988899771288899962146888986303457851258899999
Pred: HHHHHHHHHHHHHCCCHHHHHHHHCCCCCCCCCHHHHHHHHHHHCCCCCCCHHHHHHHHH
AA: RKASLALIRKMIHFCSEALLKEVCDSDVGHNLPTILVEITATVLDQEDDDDGHLLALQII
550 560 570 580 590 600
Conf: 999820260578988852035675200499999942001598877884121776307985
Pred: HHHHHCCCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHCCCC
AA: RDLVDKGGDIFLDQLARLGVISKVSTLAGPSSDDENEEESKPEKEDEPQEDAKELQQGKP
610 620 630 640 650 660
Conf: 442122564067740460210220224799860688742720144036999999998622
Pred: CCCCCCEEEECCCCCEEECCCHHHHCCCCCCCEEEEEECCCCEEEECCCCCCCCCCCCCC
AA: YHWRDWSIIRGRDCLYIWSDAAALELSNGSNGWFRFILDGKLATMYSSGSPEGGSDSSES
670 680 690 700 710 720
Conf: 379999999850333899986563258998813441034572454407996798542125
Pred: HHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCEEECCEEEEECCCCEEEEECCCCCCEEE
AA: RSEFLEKLQRARGQVKPSTSSQPILSAPGPTKLTVGNWSLTCLKEGEIAIHNSDGQQATI
730 740 750 760 770 780
Conf: 213589947860899734200024445432111046776644035799899999999999
Pred: ECCCCCCCEEECCCCCCCCCEECCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHH
AA: LKEDLPGFVFESNRGTKHSFTAETSLGSEFVTGWTGKRGRKLKSKLEKTKQKVRTMARDL
790 800 810 820 830 840
Conf: 999987310166258999999999999999984032355542208999999999999973
Pred: HHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHH
AA: YDDHFKAVESMPRGVVVTLRNIATQLESSWELHTNRQCIESENTWRDLMKTALENLIVLL
850 860 870 880 890 900
Conf: 269942420022000899999840359895122324567999999997404588888770
Pred: CCCCCCCCCCCCCHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHCCCCCCCCCH
AA: KDENTISPYEMCSSGLVQALLTVLNNSMDLDMKQDCSQLVERINVFKTAFSENEDDESRP
910 920 930 940 950 960
Conf: 999999999987600136743346999977335563442000013888752023558721
Pred: HHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHEECCCCCCCCCCCCCCCCC
AA: AVALIRKLIAVLESIERLPLHLYDTPGSTYNLQILTRRLRFRLERAPGETALIDRTGRML
970 980 990 1000 1010 1020
Conf: 344330079999999997774640368872344554025984589940356989439998
Pred: CCCCCCHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHCCCCCEEEEEECCCCCCCEEEEE
AA: KMEPLATVESLEQYLLKMVAKQWYDFDRSSFVFVRKLREGQNFIFRHQHDFDENGIIYWI
1030 1040 1050 1060 1070 1080
Conf: 157764322238531003999836998788887210113688764445478886338993
Pred: CCCCCCCCCCCCCCCEEEEEEECCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCEEEEE
AA: GTNAKTAYEWVNPAAYGLVVVTSSEGRNLPYGRLEDILSRDNSALNCHSNDDKNAWFAID
1090 1100 1110 1120 1130 1140
Conf: 062471574100011264843223306787238874136857406687789999640025
Pred: CCCEEECCCCCCCCCCCCCCCCCCCEEEEEECCCCCCEEEEEECCCCCCCCCCCCCCCCC
AA: LGLWVIPSAYTLRHARGYGRSALRNWVFQVSKDGQNWTSLYTHVDDCSLNEPGSTATWPL
1150 1160 1170 1180 1190 1200
Conf: 899987886305775224868889813201245300117999980501234568999999
Pred: CCCCCCCCCCEEEEEECCCCCCCCCCEECCCCCCCCCEEEEEEECCCHHHHHHHHHHHHH
AA: DPPKDEKQGWRHVRIKQMGKNASGQTHYLSLSGFELYGTVNGVCEDQLGKAAKEAEANLR
1210 1220 1230 1240 1250 1260
Conf: 987688764211047886176314540037989999996073034476057320899875
Pred: HHHHHHHHHHHHCCCCCCEEEECCCCCCCCCCCCCCCCCCEECCCCCCCEEEEECCCCCC
AA: RQRRLVRSQVLKYMVPGARVIRGLDWKWRDQDGSPQGEGTVTGELHNGWIDVTWDAGGSN
1270 1280 1290 1300 1310 1320
Conf: 545577997543347899999888888887665576545665545899997754457776
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: SYRMGAEGKFDLKLAPGYDPDTVASPKPVSSTVSGTTQSWSSLVKNNCPDKTSAAAGSSS
1330 1340 1350 1360 1370 1380
Conf: 444577776544445567787511222100101234567777778031145776674223
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHCCCCCCCCCCCCCCEECCCCCCCCCCCC
AA: RKGSSSSVCSVASSSDISLGSTKTERRSEIVMEHSIVSGADVHEPIVVLSSAENVPQTEV
1390 1400 1410 1420 1430 1440
Conf: 588865665322235864433557999888888988764446434569988873111122
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHH
AA: GSSSSASTSTLTAETGSENAERKLGPDSSVRTPGESSAISMGIVSVSSPDVSSVSELTNK
1450 1460 1470 1480 1490 1500
Conf: 210169987544455665643358888988888889986545640022221122210124
Pred: HHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCC
AA: EAASQRPLSSSASNRLSVSSLLAAGAPMSSSASVPNLSSRETSSLESFVRRVANIARTNA
1510 1520 1530 1540 1550 1560
Conf: 688887767788987766667454444553445678988899976543457897667887
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: TNNMNLSRSSSDNNTNTLGRNVMSTATSPLMGAQSFPNLTTPGTTSTVTMSTSSVTSSSN
1570 1580 1590 1600 1610 1620
Conf: 555511002365655222234444688887564653335778852044432222369999
Pred: CCCCCEECCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHCCCCCCCCCCCCCCC
AA: VATATTVLSVGQSLSNTLTTSLTSTSSESDTGQEAEYSLYDFLDSCRASTLLAELDDDED
1630 1640 1650 1660 1670 1680
Conf: 999998667675785202458999973177421125988866644435889998889998
Pred: CCCCCCCCCCCCCCCCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: LPEPDEEDDENEDDNQEDQEYEEVMILRRPSLQRRAGSRSDVTHHAVTSQLPQVPAGAGS
1690 1700 1710 1720 1730 1740
Conf: 877521000003799986657503454012345545779999998877641000299999
Pred: CCCCCCCCCCCCCCCCCCCCCCHHHHHHCCCCCCCCCCCCCCCCCCCCCCCEEECCCCCC
AA: RPIGEQEEEEYETKGGRRRTWDDDYVLKRQFSALVPAFDPRPGRTNVQQTTDLEIPPPGT
1750 1760 1770 1780 1790 1800
Conf: 997432110269998389997508999964137427899861489999997212699854
Pred: CCCCCCCCCCCCCCCCEEEEEECCCCCCCCCEEEECCCCCCHHHHHHHHHHHHCCCCCCC
AA: PHSELLEEVECTPSPRLALTLKVTGLGTTREVELPLTNFRSTIFYYVQKLLQLSCNGNVK
1810 1820 1830 1840 1850 1860
Conf: 322212335708998235789720002698888841113220278899995114555012
Pred: CCCCCCCCCCEEEEEECCCCCCCCCCCCCCCCCCCHHHHHHCCCCCCCCCCCHHHHHHCC
AA: SDKLRRIWEPTYTIMYREMKDSDKEKENGKMGCWSIEHVEQYLGTDELPKNDLITYLQKN
1870 1880 1890 1900 1910 1920
Conf: 027776115676654223345660267999885310278889988865433434434446
Pred: HHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC
AA: ADAAFLRHWKLTGTNKSIRKNRNCSQLIAAYKDFCEHGTKSGLNQGAISTLQSSDILNLT
1930 1940 1950 1960 1970 1980
Conf: 567555579999987687899999999884389887775457854454345954111554
Pred: CCCCCCCCCCCCCCCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCHHHHH
AA: KEQPQAKAGNGQNSCGVEDVLQLLRILYIVASDPYSRISQEDGDEQLQFTFPPDEFTSKK
1990 2000 2010 2020 2030 2040
Conf: 547899883336777039990488872026984332134320266202575532785311
Pred: HHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCCCCCCCCCCEEEEEECCCCCCEEEEECC
AA: ITTKILQQIEEPLALASGALPDWCEQLTSKCPFLIPFETRQLYFTCTAFGASRAIVWLQN
2050 2060 2070 2080 2090 2100
Conf: 136677114789988789999520134334302248982369999999998604442689
Pred: CHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCEEECCCCCCHHHHHHHHHHHHHCCCCEEE
AA: RREATVERTRTTSSVRRDDPGEFRVGRLKHERVKVPRGESLMEWAENVMQIHADRKSVLE
2110 2120 2130 2140 2150 2160
Conf: 998257765777027789998775401356337705999999775324899878996266
Pred: EEEECCCCCCCCHHHHHHHHHHHHHHCCCCCEEECCCCCCCCCCCCCCCCCCCCCCCCEE
AA: VEFLGEEGTGLGPTLEFYALVAAEFQRTDLGAWLCDDNFPDDESRHVDLGGGLKPPGYYV
2170 2180 2190 2200 2210 2220
Conf: 079988789999996688889888754200010323458644567734788887058843
Pred: ECCCCCCCCCCCCCCHHHHHHHHHHHHHHHEECCCCCCCCCCCCCCCHHHHHHHCCCCCC
AA: QRSCGLFTAPFPQDSDELERITKLFHFLGIFLAKCIQDNRLVDLPISKPFFKLMCMGDIK
2230 2240 2250 2260 2270 2280
Conf: 222211003578787655743312233225466666665345443223599999999843
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: SNMSKLIYESRGDRDLHCTESQSEASTEEGHDSLSVGSFEEDSKSEFILDPPKPKPPAWF
2290 2300 2310 2320 2330 2340
Conf: 467894565320554047999999999999865028999999997442022333667899
Pred: CCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHCCCCCCCCCCCC
AA: NGILTWEDFELVNPHRARFLKEIKDLAIKRRQILSNKGLSEDEKNTKLQELVLKNPSGSG
2350 2360 2370 2380 2390 2400
Conf: 983011021366982798677761120457998744580129999999998976377999
Pred: CCCEEECCEEEEEEECCCCCCCCCEEECCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHH
AA: PPLSIEDLGLNFQFCPSSRIYGFTAVDLKPSGEDEMITMDNAEEYVDLMFDFCMHTGIQK
2410 2420 2430 2440 2450 2460
Conf: 999986136555668867899989888846078897773566521078999754479203
Pred: HHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCCCCCCHHHHHHCCCCCCCCCCCCCCH
AA: QMEAFRDGFNKVFPMEKLSSFSHEEVQMILCGNQSPSWAAEDIINYTEPKLGYTRDSPGF
2470 2480 2490 2500 2510 2520
Conf: 677887706896888763420147899899865567874059986159999998643301
Pred: HHHHHHHHCCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCEEEEEECCCCCCCCCCCCCE
AA: LRFVRVLCGMSSDERKAFLQFTTGCSTLPPGGLANLHPRLTVVRKVDATDASYPSVNTCV
2530 2540 2550 2560 2570 2580
Conf: 101489999099999999998630166689
Pred: ECCCCCCCCHHHHHHHHHHHHHHHCCCCCC
AA: HYLKLPEYSSEEIMRERLLAATMEKGFHLN
2590 2600 2610
Show Conservation Score by AL2CO
1 M 2.108
2 A 0.150
3 D 0.832
4 V 0.294
5 D 2.108
6 P 2.108
7 D 0.858
8 T 1.501
9 L 2.108
10 L 2.108
11 E 2.108
12 W 2.108
13 L 2.108
14 Q -0.112
15 M 0.447
16 G 1.508
17 Q 0.465
18 G 1.475
19 D 0.749
20 E 2.108
21 R 1.411
22 D 1.330
23 M 1.475
24 Q 2.108
25 L 1.007
26 I 0.884
27 A 2.108
28 L 2.108
29 E 2.108
30 Q 2.108
31 L 2.108
32 C 2.108
33 M 2.108
34 L 1.493
35 L 2.108
36 L 2.108
37 M 2.108
38 S 2.108
39 D 2.108
40 N 2.108
41 V 1.263
42 D 2.108
43 R 1.521
44 C 1.520
45 F 2.108
46 E 1.492
47 T -0.245
48 C 1.492
49 P 2.108
50 P 2.108
51 R 2.108
52 T 0.889
53 F 2.108
54 L 2.108
55 P 2.108
56 A 2.108
57 L 2.108
58 C 1.493
59 K 0.462
60 I 1.070
61 F 1.458
62 L 1.516
63 D 1.493
64 E 0.548
65 S -0.797
66 A 0.999
67 P 1.006
68 D 1.238
69 N 0.251
70 V 0.896
71 L 1.519
72 E 2.108
73 V 1.525
74 T 1.519
75 A 2.108
76 R 2.108
77 A 2.108
78 I 1.066
79 T 2.108
80 Y 2.108
81 Y 1.522
82 L 1.404
83 D 1.522
84 V 0.990
85 S 2.108
86 A 0.496
87 E 1.519
88 C 2.108
89 T 0.840
90 R 1.521
91 R 2.108
92 I 0.874
93 V 0.904
94 G 0.021
95 V 0.731
96 D 1.236
97 G 1.493
98 A 1.014
99 I 0.826
100 K 1.356
101 A 1.017
102 L 0.848
103 C 2.108
104 N 0.232
105 R 0.993
106 L 1.484
107 V -0.285
108 V -0.636
109 V -0.060
110 E -0.072
111 L -0.325
112 N -0.668
113 N -0.098
114 R 1.510
115 T 0.259
116 S 1.359
117 R 1.225
118 D 2.108
119 L 2.108
120 A 0.546
121 E 2.108
122 Q 2.108
123 C 2.108
124 V 0.826
125 K 1.512
126 V 0.542
127 L 1.009
128 E 1.512
129 L 0.021
130 I 0.495
131 C 1.493
132 T -0.221
133 R 2.108
134 E 2.108
135 S 0.188
136 G 0.866
137 A 1.512
138 V 1.323
139 F 0.398
140 E 1.211
141 A 0.722
142 G 0.620
143 G 1.331
144 L 1.497
145 N -0.323
146 C 0.417
147 V -0.084
148 L 0.935
149 T -0.831
150 F 0.856
151 I 1.290
152 R 0.198
153 D 0.130
154 S -0.754
155 G 0.640
156 H -0.767
157 L -0.877
158 V 0.536
159 H 1.512
160 K 0.897
161 D 2.108
162 T 1.497
163 L 0.991
164 H 0.992
165 S 2.108
166 A 0.874
167 M 2.108
168 A -0.033
169 V 1.489
170 V 1.489
171 S 0.866
172 R 1.489
173 L 1.384
174 C 2.108
175 G 0.184
176 K 2.108
177 M 0.833
178 E 1.489
179 P 1.055
180 Q -0.662
181 D -0.114
182 S -0.910
183 S -0.674
184 L 0.758
185 E -0.452
186 I -0.771
187 C 0.939
188 V 0.900
189 E -0.114
190 S 0.888
191 L 2.108
192 S 1.523
193 S -0.719
194 L 1.042
195 L 1.523
196 K -0.491
197 H 0.649
198 E 0.001
199 D 1.026
200 H -0.492
201 Q -0.904
202 V 1.511
203 S 0.545
204 D 1.375
205 G 0.225
206 A 0.675
207 L 1.049
208 R 0.848
209 C 2.108
210 F 2.108
211 A 1.042
212 S 0.910
213 L 1.241
214 A 0.777
215 D 1.506
216 R 1.506
217 F 1.357
218 T 0.134
219 R 2.108
220 R 0.764
221 G -0.429
222 V 0.112
223 D 1.088
224 P 1.049
225 A 0.539
226 P 0.660
227 L 1.536
228 A 0.670
229 K -0.892
230 H 0.192
231 G 1.394
232 L 2.108
233 T -0.214
234 E -0.687
235 E 0.095
236 L 2.108
237 L 0.746
238 S -1.150
239 R 0.886
240 M 0.594
241 A -0.790
242 A -0.117
243 A 0.131
244 G 0.160
245 G -0.715
246 T -0.857
247 V -1.005
248 S -0.519
249 G -1.004
250 P -0.774
251 S -0.770
252 S -0.765
253 A -1.038
254 C -1.051
255 K -0.825
256 P -0.646
257 G -0.810
258 R -1.161
259 S -0.877
260 T -0.625
261 T -0.902
262 G -1.089
263 A -0.979
264 P -0.883
265 S -1.056
266 T -0.894
267 T -0.483
268 A -0.807
269 D -0.702
270 S -0.882
271 K -0.735
272 L -0.987
273 S -0.579
274 N -0.498
275 Q -0.554
276 V 0.286
277 S 0.256
278 T 0.588
279 I 0.424
280 V 0.863
281 S 0.686
282 L 1.535
283 L 2.108
284 S 0.348
285 T 0.358
286 L 1.539
287 C 0.945
288 R 2.108
289 G 1.511
290 S 2.108
291 P -0.222
292 V -0.888
293 V 0.846
294 T 0.667
295 H 0.028
296 D -0.147
297 L 0.506
298 L 0.616
299 R 1.036
300 S 0.547
301 E -0.874
302 L 1.045
303 P -0.453
304 D -0.370
305 S 0.596
306 I 0.452
307 E 0.725
308 S -0.739
309 A 0.293
310 L 0.417
311 Q -0.612
312 G 0.049
313 D 0.571
314 E 0.987
315 R 1.527
316 C 0.314
317 V -0.247
318 L 1.036
319 D 0.889
320 T -0.413
321 M 1.058
322 R 1.057
323 L 0.614
324 V -0.166
325 D 1.527
326 L 0.685
327 L 0.893
328 L 0.621
329 V 0.201
330 L 1.057
331 L 1.017
332 F 0.318
333 E 1.553
334 G 1.057
335 R 1.057
336 K -0.452
337 A 0.690
338 L 0.922
339 P -0.468
340 K -0.387
341 S -0.512
342 S -0.864
343 A -0.866
344 G -0.784
345 S -0.754
346 T -0.986
347 G -0.679
348 R -0.777
349 I -0.765
350 P -0.330
351 G -0.092
352 L -1.009
353 R 0.058
354 R -0.183
355 L -0.947
356 D -0.270
357 S -0.004
358 S -0.722
359 G -0.218
360 E 0.547
361 R 1.210
362 S 0.552
363 H 1.051
364 R 1.512
365 Q 0.559
366 L 1.529
367 I 1.036
368 D 2.108
369 C 1.529
370 I 1.529
371 R 1.529
372 S 0.958
373 K 1.036
374 D 1.036
375 T 0.862
376 D 0.855
377 A 1.329
378 L 1.512
379 I -0.095
380 D -0.048
381 A 0.577
382 I 0.858
383 D 0.167
384 T 0.203
385 G 0.030
386 A -0.053
387 F 0.190
388 E 0.248
389 V 0.936
390 N 1.527
391 F 0.245
392 M 0.254
393 D 1.541
394 D 0.666
395 V 1.057
396 G 2.108
397 Q 1.057
398 T 1.057
399 L 1.541
400 L 0.671
401 N 1.057
402 W 1.342
403 A 2.108
404 S 0.888
405 A 0.928
406 F 1.057
407 G 1.541
408 T 0.640
409 Q -0.253
410 E 0.433
411 M 0.681
412 V 1.541
413 E 0.050
414 F 0.475
415 L 2.108
416 C 1.067
417 E 0.201
418 R 0.264
419 G 0.651
420 A 0.382
421 D 1.041
422 V 0.677
423 N 1.089
424 R 0.464
425 G 1.091
426 Q 0.279
427 R 0.715
428 S 0.681
429 S 1.067
430 S 0.939
431 L 2.108
432 H 1.091
433 Y 1.091
434 A 2.108
435 A 0.681
436 C 0.358
437 F 1.091
438 G 1.541
439 R 1.091
440 P 0.955
441 Q -0.295
442 V 0.880
443 A 0.550
444 K 1.404
445 T 0.712
446 L 2.108
447 L 2.108
448 R -0.182
449 H -0.245
450 G 1.421
451 A 2.108
452 N 0.960
453 P 0.964
454 D 0.923
455 L 1.067
456 R 1.111
457 D 0.698
458 E 1.548
459 D 0.617
460 G 1.078
461 K 1.067
462 T 1.067
463 P 0.603
464 L 1.076
465 D 0.985
466 K 1.067
467 A 1.067
468 R 1.111
469 E 1.548
470 R 1.067
471 G 0.682
472 H 1.067
473 S 0.028
474 E 0.946
475 V 1.067
476 V -0.410
477 A -0.419
478 I 0.620
479 L 1.548
480 Q 1.247
481 S 0.368
482 P 1.518
483 G 0.592
484 D -0.148
485 W 0.288
486 M -0.001
487 C -0.814
488 P -0.620
489 V -1.024
490 N -0.911
491 K -1.167
492 G -1.147
493 D -1.144
494 D -1.179
495 K -1.005
496 K -1.209
497 K -0.985
498 K -1.341
499 D -1.020
500 T -0.927
501 N -1.034
502 K -1.191
503 D -0.977
504 E -1.213
505 E -1.189
506 E -0.456
507 C -1.340
508 N -1.321
509 E 0.577
510 P 0.293
511 K 0.151
512 G 1.541
513 D 1.527
514 P -0.205
515 E 0.600
516 M 0.623
517 A 1.554
518 P 0.083
519 I -0.190
520 Y 1.045
521 L 0.483
522 K -0.331
523 R -0.874
524 L 1.037
525 L 0.589
526 P 1.585
527 V 0.169
528 F 1.346
529 A 0.245
530 Q -0.851
531 T -0.043
532 F 0.907
533 Q -0.463
534 Q -0.810
535 T 0.799
536 M 0.271
537 L -0.449
538 P -0.869
539 S 0.691
540 I 0.896
541 R 0.807
542 K 0.886
543 A 0.282
544 S 0.239
545 L 1.585
546 A -0.122
547 L 0.731
548 I 0.749
549 R 0.567
550 K 1.564
551 M 0.889
552 I 0.732
553 H -0.150
554 F 0.843
555 C -0.349
556 S -1.219
557 E -0.457
558 A -1.131
559 L -0.339
560 L 0.730
561 K -1.114
562 E -0.555
563 V -0.465
564 C -0.239
565 D -1.013
566 S -0.740
567 D -0.797
568 V -1.314
569 G -1.122
570 H -1.028
571 N -0.567
572 L -0.515
573 P -0.104
574 T -0.641
575 I -0.781
576 L 0.636
577 V 0.098
578 E 0.066
579 I 0.835
580 T 0.396
581 A 0.021
582 T -0.273
583 V 0.388
584 L 1.139
585 D 0.086
586 Q -0.755
587 E -0.061
588 D 0.876
589 D 0.618
590 D 0.286
591 D 0.561
592 G 0.613
593 H 0.346
594 L -0.337
595 L -0.420
596 A 0.202
597 L 1.093
598 Q -0.717
599 I 0.262
600 I 0.086
601 R -0.315
602 D 0.197
603 L 0.673
604 V 0.172
605 D -1.231
606 K 0.760
607 G -0.502
608 G -1.018
609 D 0.012
610 I -0.506
611 F 0.887
612 L 0.290
613 D -0.500
614 Q 0.045
615 L 1.311
616 A 0.482
617 R 0.385
618 L 1.116
619 G 1.575
620 V 0.534
621 I 0.072
622 S -0.565
623 K 0.292
624 V 0.926
625 S -0.719
626 T -1.018
627 L 0.864
628 A -0.222
629 G -0.664
630 P -0.877
631 S -1.032
632 S -1.054
633 D -1.082
634 D -1.413
635 E -1.212
636 N -0.996
637 E -1.205
638 E -1.449
639 E -1.185
640 S -1.222
641 K -1.107
642 P -1.160
643 E -0.934
644 K -1.342
645 E -1.107
646 D -0.741
647 E -1.389
648 P -1.369
649 Q -1.211
650 E 0.043
651 D 0.069
652 A -0.086
653 K -0.709
654 E -0.238
655 L 0.143
656 Q -0.836
657 Q -0.753
658 G 0.458
659 K -0.410
660 P 0.242
661 Y 1.569
662 H 0.022
663 W 1.101
664 R -0.059
665 D 0.335
666 W 1.101
667 S -0.357
668 I 0.411
669 I -0.037
670 R 0.666
671 G 0.740
672 R 0.419
673 D 0.163
674 C 0.925
675 L 0.629
676 Y 1.377
677 I 0.156
678 W 1.541
679 S 0.953
680 D 0.342
681 A -0.365
682 A -0.213
683 A 1.087
684 L 1.227
685 E 1.541
686 L 1.541
687 S 1.380
688 N 0.279
689 G 0.715
690 S 1.557
691 N 2.108
692 G 2.108
693 W 1.557
694 F 1.557
695 R 1.557
696 F 1.270
697 I 0.469
698 L 0.657
699 D 0.734
700 G 0.258
701 K 0.715
702 L 0.715
703 A 0.315
704 T 1.109
705 M 1.109
706 Y 0.968
707 S 0.715
708 S 0.715
709 G 1.557
710 S 0.593
711 P 0.918
712 E 1.109
713 G -0.026
714 G 0.071
715 S -0.356
716 D 0.758
717 S -0.199
718 S 0.019
719 E 1.008
720 S 0.602
721 R 0.853
722 S -0.006
723 E 0.993
724 F 1.127
725 L 0.053
726 E 0.508
727 K 1.148
728 L 0.400
729 Q -0.033
730 R 0.589
731 A 0.517
732 R 0.605
733 G -0.682
734 Q -0.533
735 V -0.181
736 K -0.657
737 P -0.777
738 S -0.490
739 T -1.086
740 S -0.866
741 S -0.147
742 Q -0.327
743 P 0.584
744 I 0.732
745 L 1.016
746 S -0.188
747 A -0.686
748 P -0.341
749 G -0.024
750 P -0.594
751 T -0.824
752 K 0.007
753 L 0.529
754 T -0.244
755 V -0.129
756 G 0.646
757 N 0.748
758 W 1.581
759 S -0.107
760 L 0.852
761 T -0.661
762 C 0.591
763 L -0.971
764 K 0.160
765 E -0.109
766 G -0.941
767 E 0.924
768 I 0.338
769 A -0.895
770 I 1.103
771 H -0.424
772 N 1.144
773 S -0.067
774 D 0.617
775 G 0.754
776 Q 0.281
777 Q 0.726
778 A -0.546
779 T 1.381
780 I 0.273
781 L 0.967
782 K 0.009
783 E 0.274
784 D 0.605
785 L -0.013
786 P -0.555
787 G 1.581
788 F 1.581
789 V 0.440
790 F 1.582
791 E 0.758
792 S 0.754
793 N 0.776
794 R 0.175
795 G 0.754
796 T 0.550
797 K 0.554
798 H 1.148
799 S -0.325
800 F 0.695
801 T 0.017
802 A 0.593
803 E 0.440
804 T 0.282
805 S -0.577
806 L 1.124
807 G -0.107
808 S -0.136
809 E 0.879
810 F 0.668
811 V -0.373
812 T -0.358
813 G 0.645
814 W 0.484
815 T -0.558
816 G -0.251
817 K -1.061
818 R -0.336
819 G -0.605
820 R -0.160
821 K -0.193
822 L -0.837
823 K -0.591
824 S -0.541
825 K 0.535
826 L -0.988
827 E 0.231
828 K -0.867
829 T -0.323
830 K 0.636
831 Q -1.122
832 K 0.615
833 V 0.930
834 R -0.582
835 T -1.133
836 M -0.566
837 A 0.948
838 R -0.454
839 D -0.171
840 L 0.813
841 Y 0.303
842 D -0.317
843 D -1.042
844 H 0.506
845 F 0.911
846 K -0.501
847 A -0.298
848 V 0.712
849 E -0.367
850 S -0.680
851 M -0.794
852 P 0.790
853 R 0.781
854 G -0.192
855 V -0.273
856 V 0.608
857 V -0.373
858 T -0.519
859 L 1.070
860 R -1.033
861 N -0.464
862 I 0.541
863 A -0.011
864 T -1.324
865 Q -0.823
866 L 0.273
867 E -0.181
868 S -1.034
869 S -0.272
870 W -0.907
871 E -0.908
872 L -0.996
873 H -0.527
874 T -1.351
875 N -1.079
876 R -1.106
877 Q -1.146
878 C -0.778
879 I -1.377
880 E -1.082
881 S -1.032
882 E -1.205
883 N -1.240
884 T -1.210
885 W -0.081
886 R -1.015
887 D -1.085
888 L -0.986
889 M 0.377
890 K -1.238
891 T -0.745
892 A 0.120
893 L 0.361
894 E -0.876
895 N -0.710
896 L 0.100
897 I -0.818
898 V -1.131
899 L 0.465
900 L 0.334
901 K -0.893
902 D 0.751
903 E 0.870
904 N -0.758
905 T -1.104
906 I 0.201
907 S 2.108
908 P 0.456
909 Y 1.212
910 E 1.582
911 M 0.074
912 C 0.075
913 S 0.242
914 S 1.587
915 G 1.567
916 L 0.930
917 V 0.802
918 Q 0.064
919 A 0.216
920 L 1.592
921 L 0.230
922 T -0.880
923 V -0.033
924 L 0.984
925 N -0.239
926 N -1.321
927 S -1.092
928 M -1.477
929 D -1.004
930 L -1.216
931 D -1.396
932 M -1.324
933 K -1.124
934 Q -1.099
935 D -1.401
936 C -1.031
937 S -0.569
938 Q -0.975
939 L -1.104
940 V -0.944
941 E -0.559
942 R 0.434
943 I -0.006
944 N -0.947
945 V -0.183
946 F 1.046
947 K -0.163
948 T -0.984
949 A -0.594
950 F 0.039
951 S -1.222
952 E -1.223
953 N -1.127
954 E -1.111
955 D -0.738
956 D -1.212
957 E -1.025
958 S -1.305
959 R -0.850
960 P -0.439
961 A -0.045
962 V -0.722
963 A -0.600
964 L 1.133
965 I 0.043
966 R -0.955
967 K 1.323
968 L 1.036
969 I 0.882
970 A 0.497
971 V 1.066
972 L 2.108
973 E 1.567
974 S 1.008
975 I 0.434
976 E 2.108
977 R 0.899
978 L 0.915
979 P 0.786
980 L -0.032
981 H -0.141
982 L -0.816
983 Y 0.437
984 D 0.569
985 T -0.669
986 P -0.257
987 G -0.069
988 S -0.256
989 T -0.586
990 Y -0.559
991 N 0.072
992 L 2.108
993 Q 1.359
994 I -0.043
995 L 2.108
996 T 0.247
997 R 1.245
998 R 1.005
999 L 0.271
1000 R 0.597
1001 F 0.587
1002 R 0.814
1003 L 0.721
1004 E 0.780
1005 R 0.270
1006 A -0.573
1007 P -0.566
1008 G -0.447
1009 E 0.994
1010 T -0.511
1011 A -0.276
1012 L 0.433
1013 I -0.495
1014 D 1.274
1015 R 0.363
1016 T 0.788
1017 G 0.789
1018 R 0.772
1019 M -0.753
1020 L 1.267
1021 K 1.554
1022 M -0.028
1023 E 2.108
1024 P 1.050
1025 L 2.108
1026 A 0.708
1027 T 0.669
1028 V 1.256
1029 E -0.961
1030 S -0.265
1031 L 0.908
1032 E -0.197
1033 Q -0.446
1034 Y 0.709
1035 L 1.407
1036 L -0.260
1037 K 1.139
1038 M 0.379
1039 V 1.006
1040 A 1.142
1041 K 0.936
1042 Q 1.567
1043 W 2.108
1044 Y 0.827
1045 D 2.108
1046 F -0.329
1047 D 0.890
1048 R 1.592
1049 S 0.032
1050 S 0.517
1051 F 0.470
1052 V -0.892
1053 F 1.217
1054 V 0.363
1055 R 0.313
1056 K -0.265
1057 L 0.035
1058 R 0.052
1059 E -0.039
1060 G -0.762
1061 Q -1.004
1062 N -1.249
1063 F -0.401
1064 I -1.157
1065 F 0.948
1066 R -0.950
1067 H 0.875
1068 Q -0.592
1069 H -0.821
1070 D 2.108
1071 F 2.108
1072 D 2.108
1073 E 0.358
1074 N 0.161
1075 G 2.108
1076 I 0.685
1077 I 0.224
1078 Y 1.482
1079 W 0.761
1080 I 0.554
1081 G 2.108
1082 T 1.318
1083 N 2.108
1084 A 0.746
1085 K 0.453
1086 T 0.150
1087 A -0.298
1088 Y -0.620
1089 E 0.778
1090 W 1.567
1091 V 0.772
1092 N 2.108
1093 P 2.108
1094 A 0.890
1095 A -0.047
1096 Y -0.206
1097 G 0.920
1098 L 0.930
1099 V 0.614
1100 V -1.007
1101 V 1.395
1102 T 0.266
1103 S 0.607
1104 S 2.108
1105 E 0.522
1106 G 0.453
1107 R 0.598
1108 N -0.575
1109 L 1.542
1110 P 1.567
1111 Y 0.935
1112 G 1.567
1113 R 0.452
1114 L 0.220
1115 E 1.567
1116 D 1.567
1117 I 0.291
1118 L 1.166
1119 S 1.586
1120 R 2.108
1121 D 0.955
1122 N -0.725
1123 S -0.473
1124 A 0.798
1125 L -0.191
1126 N 1.087
1127 C 0.650
1128 H 1.087
1129 S 1.449
1130 N 0.021
1131 D 1.520
1132 D 0.591
1133 K -0.100
1134 N 0.209
1135 A 0.864
1136 W 0.364
1137 F 0.617
1138 A 0.211
1139 I 0.300
1140 D 1.504
1141 L 1.099
1142 G 1.504
1143 L 0.167
1144 W -0.549
1145 V 0.689
1146 I -0.164
1147 P 1.504
1148 S 0.771
1149 A -0.492
1150 Y 2.108
1151 T 0.935
1152 L 1.063
1153 R 1.504
1154 H 1.126
1155 A 1.412
1156 R 0.609
1157 G 1.097
1158 Y 1.060
1159 G 0.659
1160 R 0.334
1161 S 1.270
1162 A 1.130
1163 L 1.164
1164 R 1.584
1165 N 0.751
1166 W 1.584
1167 V -0.483
1168 F 1.324
1169 Q 1.113
1170 V -0.171
1171 S 1.113
1172 K -0.149
1173 D 1.122
1174 G 0.316
1175 Q -0.635
1176 N -0.329
1177 W 0.518
1178 T -0.928
1179 S -0.044
1180 L 0.060
1181 Y -0.916
1182 T -0.522
1183 H 0.598
1184 V -0.931
1185 D -0.305
1186 D 1.045
1187 C -0.946
1188 S -0.097
1189 L 1.166
1190 N -0.910
1191 E 0.036
1192 P 0.349
1193 G 0.943
1194 S 0.718
1195 T 0.636
1196 A -0.082
1197 T 0.400
1198 W 0.692
1199 P -0.514
1200 L 0.257
1201 D -0.741
1202 P -1.160
1203 P -0.555
1204 K -0.962
1205 D -0.375
1206 E -0.367
1207 K -1.066
1208 Q -0.876
1209 G -0.334
1210 W -0.224
1211 R 1.053
1212 H 0.333
1213 V -0.189
1214 R 1.474
1215 I 0.774
1216 K -0.577
1217 Q 0.802
1218 M -0.206
1219 G 0.460
1220 K 0.290
1221 N 0.759
1222 A 0.180
1223 S 1.216
1224 G 0.327
1225 Q 0.258
1226 T -0.231
1227 H 0.144
1228 Y 0.414
1229 L -0.084
1230 S 0.837
1231 L -0.673
1232 S 0.343
1233 G 0.430
1234 F 0.169
1235 E 1.020
1236 L 0.398
1237 Y 0.728
1238 G 0.728
1239 T -1.087
1240 V 0.488
1241 N -0.778
1242 G 0.035
1243 V 0.766
1244 C -0.418
1245 E -0.146
1246 D -0.410
1247 Q -1.000 *
1248 L -0.706
1249 G -0.001
1250 K -0.794
1251 A -0.516
1252 A -0.597
1253 K -0.553
1254 E -0.507
1255 A -1.067
1256 E -0.445
1257 A -0.735
1258 N -1.093
1259 L -0.805
1260 R -0.112
1261 R -0.159
1262 Q -0.231
1263 R 0.754
1264 R 0.071
1265 L -1.199
1266 V -0.919
1267 R -0.570
1268 S -1.337
1269 Q -0.419
1270 V -0.637
1271 L -0.727
1272 K 0.414
1273 Y -0.906
1274 M -0.689
1275 V -1.072
1276 P -1.066
1277 G 0.631
1278 A -0.410
1279 R 0.112
1280 V 0.019
1281 I -0.667
1282 R 0.697
1283 G 0.440
1284 L -0.729
1285 D 1.717
1286 W 1.576
1287 K 0.036
1288 W 1.106
1289 R -0.044
1290 D 0.292
1291 Q 0.805
1292 D 0.695
1293 G 0.684
1294 S -0.748
1295 P -0.627
1296 Q -0.411
1297 G -0.021
1298 E -0.701
1299 G 1.302
1300 T -0.667
1301 V 0.123
1302 T 0.009
1303 G -0.136
1304 E -0.382
1305 L -0.217
1306 H -0.026
1307 N -0.159
1308 G 0.947
1309 W 0.774
1310 I 0.621
1311 D 0.403
1312 V 1.276
1313 T -0.692
1314 W 0.670
1315 D 0.434
1316 A -0.658
1317 G -0.099
1318 G -1.004
1319 S -0.132
1320 N 0.788
1321 S 0.142
1322 Y -0.000
1323 R -0.221
1324 M -0.612
1325 G -0.051
1326 A -0.315
1327 E -0.182
1328 G -0.542
1329 K -0.270
1330 F -0.271
1331 D 0.586
1332 L 0.132
1333 K -0.504
1334 L -0.476
1335 A -0.638
1336 P -0.536
1337 G -1.080
1338 Y -1.398
1339 D -1.058
1340 P -0.761
1341 D -0.997
1342 T -1.138
1343 V -1.395
1344 A -1.328
1345 S -1.151
1346 P -1.284
1347 K -1.334
1348 P -0.897
1349 V -1.317
1350 S -1.118
1351 S -0.891
1352 T -1.157
1353 V -1.392
1354 S -1.174
1355 G -1.075
1356 T -0.844
1357 T -1.064
1358 Q -1.052
1359 S -1.012
1360 W -1.577
1361 S -0.838
1362 S -1.185
1363 L -0.882
1364 V -1.109
1365 K -0.925
1366 N -1.094
1367 N -1.189
1368 C -1.138
1369 P -0.948
1370 D -0.954
1371 K -1.294
1372 T -1.273
1373 S -1.219
1374 A -1.202
1375 A -1.254
1376 A -1.213
1377 G -1.220
1378 S -0.902
1379 S -1.261
1380 S -1.188
1381 R -1.523
1382 K -0.842
1383 G -1.064
1384 S -1.052
1385 S -0.969
1386 S -1.234
1387 S -1.223
1388 V -0.975
1389 C -1.426
1390 S -0.939
1391 V -1.106
1392 A -0.749
1393 S -1.154
1394 S -1.139
1395 S -1.315
1396 D -1.347
1397 I -1.202
1398 S -1.173
1399 L -1.195
1400 G -1.038
1401 S -1.323
1402 T -0.891
1403 K -1.341
1404 T -0.989
1405 E -1.159
1406 R -1.303
1407 R -0.917
1408 S -1.086
1409 E -0.900
1410 I -1.238
1411 V -1.093
1412 M -1.135
1413 E -0.965
1414 H -1.297
1415 S -1.401
1416 I -1.145
1417 V -1.316
1418 S -1.099
1419 G -0.782
1420 A -1.458
1421 D -1.176
1422 V -1.205
1423 H -0.974
1424 E -0.886
1425 P -1.163
1426 I -1.346
1427 V -1.007
1428 V -0.616
1429 L -1.149
1430 S -0.996
1431 S -1.202
1432 A -1.205
1433 E -1.290
1434 N -1.274
1435 V -0.926
1436 P -1.344
1437 Q -1.440
1438 T -1.377
1439 E -1.257
1440 V -1.317
1441 G -1.327
1442 S -1.018
1443 S -1.237
1444 S -1.185
1445 S -1.050
1446 A -1.221
1447 S -1.161
1448 T -1.141
1449 S -1.199
1450 T -1.213
1451 L -1.104
1452 T -1.037
1453 A -1.325
1454 E -1.013
1455 T -1.362
1456 G -1.233
1457 S -1.154
1458 E -1.078
1459 N -1.109
1460 A -1.135
1461 E -1.499
1462 R -1.165
1463 K -1.341
1464 L -1.009
1465 G -1.273
1466 P -1.098
1467 D -1.514
1468 S -1.366
1469 S -1.480
1470 V -1.330
1471 R -1.382
1472 T -1.297
1473 P -1.277
1474 G -1.363
1475 E -1.378
1476 S -1.308
1477 S -1.302
1478 A -1.191
1479 I -1.291
1480 S -1.208
1481 M -1.351
1482 G -1.106
1483 I -1.412
1484 V -1.433
1485 S -0.992
1486 V -1.363
1487 S -1.117
1488 S -1.245
1489 P -1.199
1490 D -1.475
1491 V -1.037
1492 S -1.089
1493 S -1.346
1494 V -1.172
1495 S -1.188
1496 E -1.077
1497 L -1.044
1498 T -1.166
1499 N -1.029
1500 K -1.192
1501 E -1.279
1502 A -1.456
1503 A -1.174
1504 S -1.256
1505 Q -1.090
1506 R -1.428
1507 P -1.303
1508 L -1.040
1509 S -1.065
1510 S -1.211
1511 S -0.877
1512 A -1.119
1513 S -1.166
1514 N -1.158
1515 R -1.460
1516 L -1.321
1517 S -1.121
1518 V -1.252
1519 S -1.433
1520 S -1.212
1521 L -1.420
1522 L -1.296
1523 A -1.058
1524 A -1.415
1525 G -1.050
1526 A -1.193
1527 P -1.017
1528 M -1.189
1529 S -1.260
1530 S -1.189
1531 S -0.614
1532 A -1.094
1533 S -1.025
1534 V -0.932
1535 P -0.960
1536 N -1.014
1537 L -1.177
1538 S -1.014
1539 S -1.350
1540 R -1.365
1541 E -1.094
1542 T -1.367
1543 S -1.358
1544 S -1.090
1545 L -1.448
1546 E -1.194
1547 S -0.689
1548 F -1.196
1549 V -1.035
1550 R -0.741
1551 R -1.166
1552 V -1.225
1553 A -1.178
1554 N -0.774
1555 I -1.064
1556 A -1.130
1557 R -0.986
1558 T -1.088
1559 N -0.996
1560 A -0.819
1561 T -0.897
1562 N -0.671
1563 N -1.354
1564 M -1.131
1565 N -1.278
1566 L -0.990
1567 S -0.983
1568 R -1.425
1569 S -1.270
1570 S -1.384
1571 S -1.101
1572 D -0.979
1573 N -0.991
1574 N -1.004
1575 T -1.279
1576 N -0.878
1577 T -1.322
1578 L -0.940
1579 G -0.949
1580 R -1.206
1581 N -1.240
1582 V -0.989
1583 M -1.017
1584 S -1.009
1585 T -1.180
1586 A -0.839
1587 T -1.236
1588 S -1.079
1589 P -1.227
1590 L -0.977
1591 M -1.268
1592 G -1.067
1593 A -1.276
1594 Q -1.005
1595 S -0.936
1596 F -1.268
1597 P -0.884
1598 N -1.101
1599 L -0.984
1600 T -1.035
1601 T -0.884
1602 P -1.245
1603 G -1.056
1604 T -0.994
1605 T -1.152
1606 S -0.951
1607 T -1.009
1608 V -1.257
1609 T -1.075
1610 M -1.251
1611 S -1.140
1612 T -1.259
1613 S -0.347
1614 S -1.064
1615 V -0.855
1616 T -1.230
1617 S -0.913
1618 S -0.951
1619 S -0.961
1620 N -0.883
1621 V -1.370
1622 A -1.064
1623 T -1.063
1624 A -0.909
1625 T -1.012
1626 T -0.706
1627 V -1.336
1628 L -1.129
1629 S -0.972
1630 V -0.788
1631 G -0.577
1632 Q -0.971
1633 S -1.033
1634 L -0.963
1635 S -1.005
1636 N -0.882
1637 T -0.857
1638 L -0.584
1639 T -0.379
1640 T -0.999
1641 S -0.580
1642 L -0.392
1643 T -1.096
1644 S -0.516
1645 T -0.804
1646 S -0.480
1647 S -1.035
1648 E -1.029
1649 S -0.715
1650 D -0.709
1651 T -1.223
1652 G -1.076
1653 Q -0.982
1654 E -1.162
1655 A -0.302
1656 E -0.787
1657 Y -1.390
1658 S -0.525
1659 L -0.836
1660 Y -0.400
1661 D -0.356
1662 F -0.528
1663 L -0.693
1664 D -0.355
1665 S -0.582
1666 C -0.518
1667 R -1.006
1668 A -0.668
1669 S -0.838
1670 T -0.538
1671 L -0.625
1672 L -0.763
1673 A -0.707
1674 E -0.835
1675 L -0.600
1676 D -0.369
1677 D -0.749
1678 D -0.681
1679 E -0.762
1680 D -0.914
1681 L -1.112
1682 P -0.921
1683 E -1.216
1684 P -1.198
1685 D -1.038
1686 E -0.639
1687 E -0.430
1688 D -0.416
1689 D -0.661
1690 E -0.591
1691 N -0.669
1692 E -0.339
1693 D -0.773
1694 D -1.098
1695 N -0.396
1696 Q -0.600
1697 E -0.846
1698 D -0.991
1699 Q -1.092
1700 E -0.869
1701 Y -1.093
1702 E -1.090
1703 E -0.922
1704 V -1.044
1705 M -0.640
1706 I -0.835
1707 L -1.099
1708 R -0.703
1709 R -0.890
1710 P -1.045
1711 S -0.950
1712 L -1.297
1713 Q -0.997
1714 R -1.139
1715 R -1.007
1716 A -1.199
1717 G -0.922
1718 S -0.534
1719 R -1.178
1720 S -0.607
1721 D -1.052
1722 V -0.970
1723 T -1.081
1724 H -1.258
1725 H -1.261
1726 A -1.019
1727 V -0.793
1728 T -0.897
1729 S -1.012
1730 Q -1.119
1731 L -0.758
1732 P -0.512
1733 Q -1.039
1734 V -0.586
1735 P -0.410
1736 A -0.821
1737 G -0.784
1738 A -0.580
1739 G -0.420
1740 S -0.905
1741 R -0.498
1742 P -1.079
1743 I -0.793
1744 G -0.954
1745 E -0.946
1746 Q -1.373
1747 E -1.546
1748 E -0.803
1749 E -0.703
1750 E -0.766
1751 Y -1.080
1752 E -0.980
1753 T -0.823
1754 K -0.782
1755 G -0.565
1756 G -0.597
1757 R -0.595
1758 R -0.760
1759 R -0.614
1760 T -0.405
1761 W -0.352
1762 D -0.146
1763 D -0.258
1764 D -0.255
1765 Y -0.669
1766 V -0.488
1767 L 0.174
1768 K -0.011
1769 R -1.000 *
1770 Q -0.561
1771 F -0.187
1772 S -0.419
1773 A -1.000 *
1774 L -1.000 *
1775 V 0.429
1776 P -1.000 *
1777 A -1.000 *
1778 F -1.000 *
1779 D -1.000 *
1780 P -1.000 *
1781 R -1.000 *
1782 P 0.284
1783 G -1.000 *
1784 R -1.000 *
1785 T -0.065
1786 N 0.130
1787 V -0.677
1788 Q -0.473
1789 Q 0.009
1790 T 0.564
1791 T -1.000 *
1792 D -1.000 *
1793 L -1.000 *
1794 E -1.000 *
1795 I -1.000 *
1796 P -1.000 *
1797 P -1.000 *
1798 P -1.000 *
1799 G -1.000 *
1800 T -0.686
1801 P -1.000 *
1802 H -0.683
1803 S -1.017
1804 E -0.720
1805 L -1.003
1806 L -0.991
1807 E -0.794
1808 E -0.994
1809 V -0.879
1810 E -0.867
1811 C -0.853
1812 T -1.025
1813 P -0.936
1814 S -0.857
1815 P 0.267
1816 R -1.087
1817 L 0.465
1818 A -0.809
1819 L 0.728
1820 T -1.050
1821 L 0.148
1822 K -0.125
1823 V 0.082
1824 T -0.422
1825 G 0.008
1826 L -0.716
1827 G -0.684
1828 T -0.299
1829 T -0.891
1830 R -0.529
1831 E -0.076
1832 V -0.477
1833 E -0.519
1834 L -0.415
1835 P -0.640
1836 L 0.063
1837 T -0.896
1838 N -0.254
1839 F -1.095
1840 R -0.942
1841 S -0.909
1842 T -0.245
1843 I 0.166
1844 F 0.657
1845 Y -0.733
1846 Y -0.704
1847 V 0.301
1848 Q 0.903
1849 K -0.947
1850 L 1.029
1851 L -0.612
1852 Q -0.843
1853 L -1.047
1854 S -1.231
1855 C -1.307
1856 N -1.286
1857 G 0.019
1858 N -0.791
1859 V -1.000 *
1860 K -0.553
1861 S -0.563
1862 D 0.050
1863 K -0.146
1864 L -0.438
1865 R 0.381
1866 R 0.395
1867 I -0.199
1868 W 1.000
1869 E 0.218
1870 P -0.064
1871 T 0.241
1872 Y 0.287
1873 T 0.265
1874 I 0.554
1875 M -0.985
1876 Y -0.014
1877 R -0.439
1878 E 0.066
1879 M -0.739
1880 K -0.860
1881 D -0.666
1882 S -1.096
1883 D -0.595
1884 K -1.319
1885 E -1.000 *
1886 K -1.000 *
1887 E -1.000 *
1888 N -1.000 *
1889 G -1.000 *
1890 K -1.000 *
1891 M -1.000 *
1892 G -1.000 *
1893 C -1.000 *
1894 W -1.000 *
1895 S -1.000 *
1896 I -1.000 *
1897 E -1.000 *
1898 H -1.000 *
1899 V -1.000 *
1900 E -1.000 *
1901 Q -1.000 *
1902 Y -1.000 *
1903 L -1.000 *
1904 G -1.000 *
1905 T -1.000 *
1906 D -1.000 *
1907 E -1.000 *
1908 L -1.000 *
1909 P -1.000 *
1910 K -1.000 *
1911 N -1.000 *
1912 D -1.000 *
1913 L -1.000 *
1914 I -1.000 *
1915 T -1.000 *
1916 Y -1.000 *
1917 L -1.000 *
1918 Q -1.000 *
1919 K -1.000 *
1920 N -1.000 *
1921 A -1.000 *
1922 D -1.000 *
1923 A -1.000 *
1924 A -1.000 *
1925 F -1.000 *
1926 L -1.000 *
1927 R -1.000 *
1928 H -1.000 *
1929 W -1.000 *
1930 K -1.000 *
1931 L -1.000 *
1932 T -1.000 *
1933 G -1.000 *
1934 T -1.000 *
1935 N -1.000 *
1936 K -1.000 *
1937 S -1.000 *
1938 I -1.000 *
1939 R -1.000 *
1940 K -1.000 *
1941 N -1.000 *
1942 R -1.000 *
1943 N -1.000 *
1944 C -1.000 *
1945 S -1.000 *
1946 Q -1.000 *
1947 L -1.000 *
1948 I -1.000 *
1949 A -1.000 *
1950 A -1.000 *
1951 Y -1.000 *
1952 K -1.000 *
1953 D -1.000 *
1954 F -1.000 *
1955 C -1.000 *
1956 E -1.000 *
1957 H -1.000 *
1958 G -1.000 *
1959 T -1.000 *
1960 K -1.000 *
1961 S -1.000 *
1962 G -1.000 *
1963 L -1.000 *
1964 N -1.000 *
1965 Q -1.000 *
1966 G -1.000 *
1967 A -1.000 *
1968 I -1.000 *
1969 S -1.000 *
1970 T -1.000 *
1971 L -1.000 *
1972 Q -1.000 *
1973 S -1.000 *
1974 S -1.000 *
1975 D -1.000 *
1976 I -1.000 *
1977 L -1.000 *
1978 N -1.000 *
1979 L -1.000 *
1980 T -1.000 *
1981 K -1.000 *
1982 E -1.000 *
1983 Q -1.000 *
1984 P -1.000 *
1985 Q -1.000 *
1986 A -1.000 *
1987 K -1.000 *
1988 A -1.000 *
1989 G -1.000 *
1990 N -1.000 *
1991 G -1.000 *
1992 Q -1.000 *
1993 N -1.000 *
1994 S -1.000 *
1995 C -1.000 *
1996 G -1.000 *
1997 V -1.000 *
1998 E -1.000 *
1999 D -1.000 *
2000 V -1.000 *
2001 L -1.000 *
2002 Q -1.000 *
2003 L -1.000 *
2004 L -1.000 *
2005 R -1.000 *
2006 I -1.000 *
2007 L -1.000 *
2008 Y -1.000 *
2009 I -1.000 *
2010 V -1.000 *
2011 A -1.000 *
2012 S -1.000 *
2013 D -1.000 *
2014 P -1.000 *
2015 Y -1.000 *
2016 S -1.000 *
2017 R -1.000 *
2018 I -1.000 *
2019 S -1.000 *
2020 Q -1.000 *
2021 E -1.000 *
2022 D -1.000 *
2023 G -1.000 *
2024 D -1.000 *
2025 E -1.000 *
2026 Q -1.000 *
2027 L -1.000 *
2028 Q -1.000 *
2029 F -1.000 *
2030 T -1.000 *
2031 F -1.000 *
2032 P -1.000 *
2033 P -1.000 *
2034 D -1.000 *
2035 E -1.000 *
2036 F -1.000 *
2037 T -1.000 *
2038 S -1.000 *
2039 K -1.000 *
2040 K -1.000 *
2041 I -1.000 *
2042 T -1.000 *
2043 T -1.000 *
2044 K -1.000 *
2045 I -1.000 *
2046 L -1.000 *
2047 Q -1.000 *
2048 Q -1.000 *
2049 I -1.000 *
2050 E -1.000 *
2051 E -1.000 *
2052 P -1.000 *
2053 L -1.000 *
2054 A -1.000 *
2055 L -1.000 *
2056 A -1.000 *
2057 S -1.000 *
2058 G -1.000 *
2059 A -1.000 *
2060 L -1.000 *
2061 P -1.000 *
2062 D -1.000 *
2063 W -1.000 *
2064 C -1.000 *
2065 E -1.000 *
2066 Q -1.000 *
2067 L -1.000 *
2068 T -1.000 *
2069 S -1.000 *
2070 K -1.000 *
2071 C -1.000 *
2072 P -1.000 *
2073 F -1.000 *
2074 L -1.000 *
2075 I -1.000 *
2076 P -1.000 *
2077 F -1.000 *
2078 E -1.000 *
2079 T -1.000 *
2080 R -1.000 *
2081 Q -1.000 *
2082 L -1.000 *
2083 Y -1.000 *
2084 F -1.000 *
2085 T -1.000 *
2086 C -1.000 *
2087 T -1.000 *
2088 A -1.000 *
2089 F -1.000 *
2090 G -1.000 *
2091 A -1.000 *
2092 S -1.000 *
2093 R -1.000 *
2094 A -1.000 *
2095 I -1.000 *
2096 V -1.000 *
2097 W -1.000 *
2098 L -1.000 *
2099 Q -1.000 *
2100 N -1.000 *
2101 R -1.000 *
2102 R -1.000 *
2103 E -1.000 *
2104 A -1.000 *
2105 T -1.000 *
2106 V -1.000 *
2107 E -1.000 *
2108 R -1.000 *
2109 T -1.000 *
2110 R -1.000 *
2111 T -1.000 *
2112 T -1.000 *
2113 S -1.000 *
2114 S -1.000 *
2115 V -1.000 *
2116 R -1.000 *
2117 R -1.000 *
2118 D -1.000 *
2119 D -1.000 *
2120 P -1.000 *
2121 G -1.000 *
2122 E -1.000 *
2123 F -1.000 *
2124 R -1.000 *
2125 V -1.000 *
2126 G -1.000 *
2127 R -1.000 *
2128 L -1.000 *
2129 K -1.000 *
2130 H -1.000 *
2131 E -1.000 *
2132 R -1.000 *
2133 V -1.000 *
2134 K -1.000 *
2135 V -1.000 *
2136 P -1.000 *
2137 R -1.000 *
2138 G -1.000 *
2139 E -1.000 *
2140 S -1.000 *
2141 L -1.000 *
2142 M -1.000 *
2143 E -1.000 *
2144 W -1.000 *
2145 A -1.000 *
2146 E -1.000 *
2147 N -1.000 *
2148 V -1.000 *
2149 M -1.000 *
2150 Q -1.000 *
2151 I -1.000 *
2152 H -1.000 *
2153 A -1.000 *
2154 D -1.000 *
2155 R -1.000 *
2156 K -1.000 *
2157 S -1.000 *
2158 V -1.000 *
2159 L -1.000 *
2160 E -1.000 *
2161 V -1.000 *
2162 E -1.000 *
2163 F -1.000 *
2164 L -1.000 *
2165 G -1.000 *
2166 E -1.000 *
2167 E -1.000 *
2168 G -1.000 *
2169 T -1.000 *
2170 G -1.000 *
2171 L -1.000 *
2172 G -1.000 *
2173 P -1.000 *
2174 T -1.000 *
2175 L -1.000 *
2176 E -1.000 *
2177 F -1.000 *
2178 Y -1.000 *
2179 A -1.000 *
2180 L -1.000 *
2181 V -1.000 *
2182 A -1.000 *
2183 A -1.000 *
2184 E -1.000 *
2185 F -1.000 *
2186 Q -1.000 *
2187 R -1.000 *
2188 T -1.000 *
2189 D -1.000 *
2190 L -1.000 *
2191 G -1.000 *
2192 A -1.000 *
2193 W -1.000 *
2194 L -1.000 *
2195 C -1.000 *
2196 D -1.000 *
2197 D -1.000 *
2198 N -1.000 *
2199 F -1.000 *
2200 P -1.000 *
2201 D -1.000 *
2202 D -1.000 *
2203 E -1.000 *
2204 S -1.000 *
2205 R -1.000 *
2206 H -1.000 *
2207 V -1.000 *
2208 D -1.000 *
2209 L -1.000 *
2210 G -1.000 *
2211 G -1.000 *
2212 G -1.000 *
2213 L -1.000 *
2214 K -1.000 *
2215 P -1.000 *
2216 P -1.000 *
2217 G -1.000 *
2218 Y -1.000 *
2219 Y -1.000 *
2220 V -1.000 *
2221 Q -1.000 *
2222 R -1.000 *
2223 S -1.000 *
2224 C -1.000 *
2225 G -1.000 *
2226 L -1.000 *
2227 F -1.000 *
2228 T -1.000 *
2229 A -1.000 *
2230 P -1.000 *
2231 F -1.000 *
2232 P -1.000 *
2233 Q -1.000 *
2234 D -1.000 *
2235 S -1.000 *
2236 D -1.000 *
2237 E -1.000 *
2238 L -1.000 *
2239 E -1.000 *
2240 R -1.000 *
2241 I -1.000 *
2242 T -1.000 *
2243 K -1.000 *
2244 L -1.000 *
2245 F -1.000 *
2246 H -1.000 *
2247 F -1.000 *
2248 L -1.000 *
2249 G -1.000 *
2250 I -1.000 *
2251 F -1.000 *
2252 L -1.000 *
2253 A -1.000 *
2254 K -1.000 *
2255 C -1.000 *
2256 I -1.000 *
2257 Q -1.000 *
2258 D -1.000 *
2259 N -1.000 *
2260 R -1.000 *
2261 L -1.000 *
2262 V -1.000 *
2263 D -1.000 *
2264 L -1.000 *
2265 P -1.000 *
2266 I -1.000 *
2267 S -1.000 *
2268 K -1.000 *
2269 P -1.000 *
2270 F -1.000 *
2271 F -1.000 *
2272 K -1.000 *
2273 L -1.000 *
2274 M -1.000 *
2275 C -1.000 *
2276 M -1.000 *
2277 G -1.000 *
2278 D -1.000 *
2279 I -1.000 *
2280 K -1.000 *
2281 S -1.000 *
2282 N -1.000 *
2283 M -1.000 *
2284 S -1.000 *
2285 K -1.000 *
2286 L -1.000 *
2287 I -1.000 *
2288 Y -1.000 *
2289 E -1.000 *
2290 S -1.000 *
2291 R -1.000 *
2292 G -1.000 *
2293 D -1.000 *
2294 R -1.000 *
2295 D -1.000 *
2296 L -1.000 *
2297 H -1.000 *
2298 C -1.000 *
2299 T -1.000 *
2300 E -1.000 *
2301 S -1.000 *
2302 Q -1.000 *
2303 S -1.000 *
2304 E -1.000 *
2305 A -1.000 *
2306 S -1.000 *
2307 T -1.000 *
2308 E -1.000 *
2309 E -1.000 *
2310 G -1.000 *
2311 H -1.000 *
2312 D -1.000 *
2313 S -1.000 *
2314 L -1.000 *
2315 S -1.000 *
2316 V -1.000 *
2317 G -1.000 *
2318 S -1.000 *
2319 F -1.000 *
2320 E -1.000 *
2321 E -1.000 *
2322 D -1.000 *
2323 S -1.000 *
2324 K -1.000 *
2325 S -1.000 *
2326 E -1.000 *
2327 F -1.000 *
2328 I -1.000 *
2329 L -1.000 *
2330 D -1.000 *
2331 P -1.000 *
2332 P -1.000 *
2333 K -1.000 *
2334 P -1.000 *
2335 K -1.000 *
2336 P -1.000 *
2337 P -1.000 *
2338 A -1.000 *
2339 W -1.000 *
2340 F -1.000 *
2341 N -1.000 *
2342 G -1.000 *
2343 I -1.000 *
2344 L -1.000 *
2345 T -1.000 *
2346 W -1.000 *
2347 E -1.000 *
2348 D -1.000 *
2349 F -1.000 *
2350 E -1.000 *
2351 L -1.000 *
2352 V -1.000 *
2353 N -1.000 *
2354 P -1.000 *
2355 H -1.000 *
2356 R -1.000 *
2357 A -1.000 *
2358 R -1.000 *
2359 F -1.000 *
2360 L -1.000 *
2361 K -1.000 *
2362 E -1.000 *
2363 I -1.000 *
2364 K -1.000 *
2365 D -1.000 *
2366 L -1.000 *
2367 A -1.000 *
2368 I -1.000 *
2369 K -1.000 *
2370 R -1.000 *
2371 R -1.000 *
2372 Q -1.000 *
2373 I -1.000 *
2374 L -1.000 *
2375 S -1.000 *
2376 N -1.000 *
2377 K -1.000 *
2378 G -1.000 *
2379 L -1.000 *
2380 S -1.000 *
2381 E -1.000 *
2382 D -1.000 *
2383 E -1.000 *
2384 K -1.000 *
2385 N -1.000 *
2386 T -1.000 *
2387 K -1.000 *
2388 L -1.000 *
2389 Q -1.000 *
2390 E -1.000 *
2391 L -1.000 *
2392 V -1.000 *
2393 L -1.000 *
2394 K -1.000 *
2395 N -1.000 *
2396 P -1.000 *
2397 S -1.000 *
2398 G -1.000 *
2399 S -1.000 *
2400 G -1.000 *
2401 P -1.000 *
2402 P -1.000 *
2403 L -1.000 *
2404 S -1.000 *
2405 I -1.000 *
2406 E -1.000 *
2407 D -1.000 *
2408 L -1.000 *
2409 G -1.000 *
2410 L -1.000 *
2411 N -1.000 *
2412 F -1.000 *
2413 Q -1.000 *
2414 F -1.000 *
2415 C -1.000 *
2416 P -1.000 *
2417 S -1.000 *
2418 S -1.000 *
2419 R -1.000 *
2420 I -1.000 *
2421 Y -1.000 *
2422 G -1.000 *
2423 F -1.000 *
2424 T -1.000 *
2425 A -1.000 *
2426 V -1.000 *
2427 D -1.000 *
2428 L -1.000 *
2429 K -1.000 *
2430 P -1.000 *
2431 S -1.000 *
2432 G -1.000 *
2433 E -1.000 *
2434 D -1.000 *
2435 E -1.000 *
2436 M -1.000 *
2437 I -1.000 *
2438 T -1.000 *
2439 M -1.000 *
2440 D -1.000 *
2441 N -1.000 *
2442 A -1.000 *
2443 E -1.000 *
2444 E -1.000 *
2445 Y -1.000 *
2446 V -1.000 *
2447 D -1.000 *
2448 L -1.000 *
2449 M -1.000 *
2450 F -1.000 *
2451 D -1.000 *
2452 F -1.000 *
2453 C -1.000 *
2454 M -1.000 *
2455 H -1.000 *
2456 T -1.000 *
2457 G -1.000 *
2458 I -1.000 *
2459 Q -1.000 *
2460 K -1.000 *
2461 Q -1.000 *
2462 M -1.000 *
2463 E -1.000 *
2464 A -1.000 *
2465 F -1.000 *
2466 R -1.000 *
2467 D -1.000 *
2468 G -1.000 *
2469 F -1.000 *
2470 N -1.000 *
2471 K -1.000 *
2472 V -1.000 *
2473 F -1.000 *
2474 P -1.000 *
2475 M -1.000 *
2476 E -1.000 *
2477 K -1.000 *
2478 L -1.000 *
2479 S -1.000 *
2480 S -1.000 *
2481 F -1.000 *
2482 S -1.000 *
2483 H -1.000 *
2484 E -1.000 *
2485 E -1.000 *
2486 V -1.000 *
2487 Q -1.000 *
2488 M -1.000 *
2489 I -1.000 *
2490 L -1.000 *
2491 C -1.000 *
2492 G -1.000 *
2493 N -1.000 *
2494 Q -1.000 *
2495 S -1.000 *
2496 P -1.000 *
2497 S -1.000 *
2498 W -1.000 *
2499 A -1.000 *
2500 A -1.000 *
2501 E -1.000 *
2502 D -1.000 *
2503 I -1.000 *
2504 I -1.000 *
2505 N -1.000 *
2506 Y -1.000 *
2507 T -1.000 *
2508 E -1.000 *
2509 P -1.000 *
2510 K -1.000 *
2511 L -1.000 *
2512 G -1.000 *
2513 Y -1.000 *
2514 T -1.000 *
2515 R -1.000 *
2516 D -1.000 *
2517 S -1.000 *
2518 P -1.000 *
2519 G -1.000 *
2520 F -1.000 *
2521 L -1.000 *
2522 R -1.000 *
2523 F -1.000 *
2524 V -1.000 *
2525 R -1.000 *
2526 V -1.000 *
2527 L -1.000 *
2528 C -1.000 *
2529 G -1.000 *
2530 M -1.000 *
2531 S -1.000 *
2532 S -1.000 *
2533 D -1.000 *
2534 E -1.000 *
2535 R -1.000 *
2536 K -1.000 *
2537 A -1.000 *
2538 F -1.000 *
2539 L -1.000 *
2540 Q -1.000 *
2541 F -1.000 *
2542 T -1.000 *
2543 T -1.000 *
2544 G -1.000 *
2545 C -1.000 *
2546 S -1.000 *
2547 T -1.000 *
2548 L -1.000 *
2549 P -1.000 *
2550 P -1.000 *
2551 G -1.000 *
2552 G -1.000 *
2553 L -1.000 *
2554 A -1.000 *
2555 N -1.000 *
2556 L -1.000 *
2557 H -1.000 *
2558 P -1.000 *
2559 R -1.000 *
2560 L -1.000 *
2561 T -1.000 *
2562 V -1.000 *
2563 V -1.000 *
2564 R -1.000 *
2565 K -1.000 *
2566 V -1.000 *
2567 D -1.000 *
2568 A -1.000 *
2569 T -1.000 *
2570 D -1.000 *
2571 A -1.000 *
2572 S -1.000 *
2573 Y -1.000 *
2574 P -1.000 *
2575 S -1.000 *
2576 V -1.000 *
2577 N -1.000 *
2578 T -1.000 *
2579 C -1.000 *
2580 V -1.000 *
2581 H -1.000 *
2582 Y -1.000 *
2583 L -1.000 *
2584 K -1.000 *
2585 L -1.000 *
2586 P -1.000 *
2587 E -1.000 *
2588 Y -1.000 *
2589 S -1.000 *
2590 S -1.000 *
2591 E -1.000 *
2592 E -1.000 *
2593 I -1.000 *
2594 M -1.000 *
2595 R -1.000 *
2596 E -1.000 *
2597 R -1.000 *
2598 L -1.000 *
2599 L -1.000 *
2600 A -1.000 *
2601 A -1.000 *
2602 T -1.000 *
2603 M -1.000 *
2604 E -1.000 *
2605 K -1.000 *
2606 G -1.000 *
2607 F -1.000 *
2608 H -1.000 *
2609 L -1.000 *
2610 N -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 334.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60MADVDPDTLL EWLQMGQGDE RDMQLIALEQ LCMLLLMSDN VDRCFETCPP RTFLPALCKI70 80 90 100 110 120FLDESAPDNV LEVTARAITY YLDVSAECTR RIVGVDGAIK ALCNRLVVVE LNNRTSRDLA130 140 150 160 170 180EQCVKVLELI CTRESGAVFE AGGLNCVLTF IRDSGHLVHK DTLHSAMAVV SRLCGKMEPQ190 200 210 220 230 240DSSLEICVES LSSLLKHEDH QVSDGALRCF ASLADRFTRR GVDPAPLAKH GLTEELLSRM250 260 270 280 290 300AAAGGTVSGP SSACKPGRST TGAPSTTADS KLSNQVSTIV SLLSTLCRGS PVVTHDLLRS310 320 330 340 350 360ELPDSIESAL QGDERCVLDT MRLVDLLLVL LFEGRKALPK SSAGSTGRIP GLRRLDSSGE370 380 390 400 410 420RSHRQLIDCI RSKDTDALID AIDTGAFEVN FMDDVGQTLL NWASAFGTQE MVEFLCERGA430 440 450 460 470 480DVNRGQRSSS LHYAACFGRP QVAKTLLRHG ANPDLRDEDG KTPLDKARER GHSEVVAILQ490 500 510 520 530 540SPGDWMCPVN KGDDKKKKDT NKDEEECNEP KGDPEMAPIY LKRLLPVFAQ TFQQTMLPSI550 560 570 580 590 600RKASLALIRK MIHFCSEALL KEVCDSDVGH NLPTILVEIT ATVLDQEDDD DGHLLALQII610 620 630 640 650 660RDLVDKGGDI FLDQLARLGV ISKVSTLAGP SSDDENEEES KPEKEDEPQE DAKELQQGKP670 680 690 700 710 720YHWRDWSIIR GRDCLYIWSD AAALELSNGS NGWFRFILDG KLATMYSSGS PEGGSDSSES730 740 750 760 770 780RSEFLEKLQR ARGQVKPSTS SQPILSAPGP TKLTVGNWSL TCLKEGEIAI HNSDGQQATI790 800 810 820 830 840LKEDLPGFVF ESNRGTKHSF TAETSLGSEF VTGWTGKRGR KLKSKLEKTK QKVRTMARDL850 860 870 880 890 900YDDHFKAVES MPRGVVVTLR NIATQLESSW ELHTNRQCIE SENTWRDLMK TALENLIVLL910 920 930 940 950 960KDENTISPYE MCSSGLVQAL LTVLNNSMDL DMKQDCSQLV ERINVFKTAF SENEDDESRP970 980 990 1000 1010 1020AVALIRKLIA VLESIERLPL HLYDTPGSTY NLQILTRRLR FRLERAPGET ALIDRTGRML1030 1040 1050 1060 1070 1080KMEPLATVES LEQYLLKMVA KQWYDFDRSS FVFVRKLREG QNFIFRHQHD FDENGIIYWI1090 1100 1110 1120 1130 1140GTNAKTAYEW VNPAAYGLVV VTSSEGRNLP YGRLEDILSR DNSALNCHSN DDKNAWFAID1150 1160 1170 1180 1190 1200LGLWVIPSAY TLRHARGYGR SALRNWVFQV SKDGQNWTSL YTHVDDCSLN EPGSTATWPL1210 1220 1230 1240 1250 1260DPPKDEKQGW RHVRIKQMGK NASGQTHYLS LSGFELYGTV NGVCEDQLGK AAKEAEANLR1270 1280 1290 1300 1310 1320RQRRLVRSQV LKYMVPGARV IRGLDWKWRD QDGSPQGEGT VTGELHNGWI DVTWDAGGSN1330 1340 1350 1360 1370 1380SYRMGAEGKF DLKLAPGYDP DTVASPKPVS STVSGTTQSW SSLVKNNCPD KTSAAAGSSS1390 1400 1410 1420 1430 1440RKGSSSSVCS VASSSDISLG STKTERRSEI VMEHSIVSGA DVHEPIVVLS SAENVPQTEV1450 1460 1470 1480 1490 1500GSSSSASTST LTAETGSENA ERKLGPDSSV RTPGESSAIS MGIVSVSSPD VSSVSELTNK1510 1520 1530 1540 1550 1560EAASQRPLSS SASNRLSVSS LLAAGAPMSS SASVPNLSSR ETSSLESFVR RVANIARTNA1570 1580 1590 1600 1610 1620TNNMNLSRSS SDNNTNTLGR NVMSTATSPL MGAQSFPNLT TPGTTSTVTM STSSVTSSSN1630 1640 1650 1660 1670 1680VATATTVLSV GQSLSNTLTT SLTSTSSESD TGQEAEYSLY DFLDSCRAST LLAELDDDED1690 1700 1710 1720 1730 1740LPEPDEEDDE NEDDNQEDQE YEEVMILRRP SLQRRAGSRS DVTHHAVTSQ LPQVPAGAGS1750 1760 1770 1780 1790 1800RPIGEQEEEE YETKGGRRRT WDDDYVLKRQ FSALVPAFDP RPGRTNVQQT TDLEIPPPGT1810 1820 1830 1840 1850 1860PHSELLEEVE CTPSPRLALT LKVTGLGTTR EVELPLTNFR STIFYYVQKL LQLSCNGNVK1870 1880 1890 1900 1910 1920SDKLRRIWEP TYTIMYREMK DSDKEKENGK MGCWSIEHVE QYLGTDELPK NDLITYLQKN1930 1940 1950 1960 1970 1980ADAAFLRHWK LTGTNKSIRK NRNCSQLIAA YKDFCEHGTK SGLNQGAIST LQSSDILNLT1990 2000 2010 2020 2030 2040KEQPQAKAGN GQNSCGVEDV LQLLRILYIV ASDPYSRISQ EDGDEQLQFT FPPDEFTSKK2050 2060 2070 2080 2090 2100ITTKILQQIE EPLALASGAL PDWCEQLTSK CPFLIPFETR QLYFTCTAFG ASRAIVWLQN2110 2120 2130 2140 2150 2160RREATVERTR TTSSVRRDDP GEFRVGRLKH ERVKVPRGES LMEWAENVMQ IHADRKSVLE2170 2180 2190 2200 2210 2220VEFLGEEGTG LGPTLEFYAL VAAEFQRTDL GAWLCDDNFP DDESRHVDLG GGLKPPGYYV2230 2240 2250 2260 2270 2280QRSCGLFTAP FPQDSDELER ITKLFHFLGI FLAKCIQDNR LVDLPISKPF FKLMCMGDIK2290 2300 2310 2320 2330 2340SNMSKLIYES RGDRDLHCTE SQSEASTEEG HDSLSVGSFE EDSKSEFILD PPKPKPPAWF2350 2360 2370 2380 2390 2400NGILTWEDFE LVNPHRARFL KEIKDLAIKR RQILSNKGLS EDEKNTKLQE LVLKNPSGSG2410 2420 2430 2440 2450 2460PPLSIEDLGL NFQFCPSSRI YGFTAVDLKP SGEDEMITMD NAEEYVDLMF DFCMHTGIQK2470 2480 2490 2500 2510 2520QMEAFRDGFN KVFPMEKLSS FSHEEVQMIL CGNQSPSWAA EDIINYTEPK LGYTRDSPGF2530 2540 2550 2560 2570 2580LRFVRVLCGM SSDERKAFLQ FTTGCSTLPP GGLANLHPRL TVVRKVDATD ASYPSVNTCV2590 2600 2610HYLKLPEYSS EEIMRERLLA ATMEKGFHLN
3D Structures in PDB
Comments
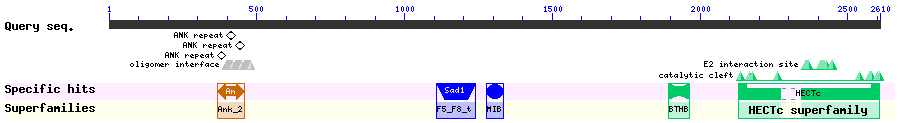
The HECT domain found in E3 ubiquitin ligase 1, HECTD1, interacts with the transcription factor SNAIL and impacts the stability of SNAIL by way of ubiquitination. HECTD1 contains eight putative NLSs and four putative NESs and its translocation between the nucleus and cytoplasm is regulated by epidermal growth factor (EGF). Experimental LMB treatment of HECTD1 resulted in a loss of nuclear export as well as an observed loss of SNAIL expression. Putative NESs were identified as amino acids 33-35, 314-333, and 599-602. The putative NES 599-602 is predicted to be in the ANK 4 repeat.
References
[1]. "HECTD1 Regulates the Expression of SNAIL: Implications for Epithelial-mesenchymal Transition"
Wang X, De Geyter C, Jia Z, Peng Y, Zhang H. (2020) Int J Oncol, 56(5):1186-1198 PubMed
Wang X, De Geyter C, Jia Z, Peng Y, Zhang H. (2020) Int J Oncol, 56(5):1186-1198 PubMed
User Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.