Show FASTA Format
>tr|C7GRB6|C7GRB6_YEAS2 Rna1p OS=Saccharomyces cerevisiae (strain JAY291) OX=574961 GN=RNA1 PE=4 SV=1
MATLHFVPQHEEEQVYSISGKALKLTTSDDIKPYLEELAALKTCTKLDLSGNTIGTEASE
ALAKCIAENTQVRESLVEVNFADLYTSRLVDEVVDSLKFLLPVLLKCPHLEIVNLSDNAF
GLRTIELLEDYIAHAVNIKHLILSNNGMGPFAGERIGKALFHLAQNKKAASKPFLETFIC
GRNRLENGSAVYLALGLKSHSEGLKVVKLYQNGIRPKGVATLIHYGLQYLKNLEILDLQD
NTFTKHASLILAKALPTWKDSLFELNLNDCLLKTAGSDEVFKVFTEVKFPNLHVLKFEYN
EMAQETIEVSFLPAMEKGNLPELEKLEINGNRLDEDSDALDLLQSKFDDLEVDDFEEVDS
EDEEGEDEEDEDEDEKLEEIETERLEKELLEVQVDDLAERLAETEIK
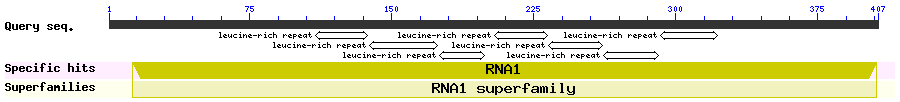
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 975334455678705767156448999327899999964399931998579744799999
Pred: CCCCCCCCCCCCCEEEEEECCCCCCCCHHCHHHHHHHHHCCCCCEEEECCCCCCCHHHHH
AA: MATLHFVPQHEEEQVYSISGKALKLTTSDDIKPYLEELAALKTCTKLDLSGNTIGTEASE
10 20 30 40 50 60
Conf: 999999407333410021355410115784147999999689771699831882998978
Pred: HHHHHHHHCHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHCCCCCCEEECCCCCC
AA: ALAKCIAENTQVRESLVEVNFADLYTSRLVDEVVDSLKFLLPVLLKCPHLEIVNLSDNAF
70 80 90 100 110 120
Conf: 868738999996229873543215899983578999999999999756439999328982
Pred: CCCCHHHHHHHHHCCCCCCEEECCCCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCEEEE
AA: GLRTIELLEDYIAHAVNIKHLILSNNGMGPFAGERIGKALFHLAQNKKAASKPFLETFIC
130 140 150 160 170 180
Conf: 577556632899999999826884399825899995789999996872199998651788
Pred: CCCCCCCCCHHHHHHHHHHCCCCCEEEECCCCCCCHHHHHHHHHHHHHCCCCCCEEECCC
AA: GRNRLENGSAVYLALGLKSHSEGLKVVKLYQNGIRPKGVATLIHYGLQYLKNLEILDLQD
190 200 210 220 230 240
Conf: 756704699999651001210246408643457889899999860499999428982478
Pred: CCCCCHHHHHHHHHCCHHHHCCCEEECCCCCCCCCCHHHHHHHHHCCCCCCCCEEECCCC
AA: NTFTKHASLILAKALPTWKDSLFELNLNDCLLKTAGSDEVFKVFTEVKFPNLHVLKFEYN
250 260 270 280 290 300
Conf: 887567754211234416999865688248879989636999999777349987777776
Pred: CCCHHHHHHHHHHHHHHCCCCCCCEEECCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCC
AA: EMAQETIEVSFLPAMEKGNLPELEKLEINGNRLDEDSDALDLLQSKFDDLEVDDFEEVDS
310 320 330 340 350 360
Conf: 66668888653101234555665555532100479999996315679
Pred: CCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHCHHHHHHHHHCCCCC
AA: EDEEGEDEEDEDEDEKLEEIETERLEKELLEVQVDDLAERLAETEIK
370 380 390 400
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 T -1.000 *
4 L -1.000 *
5 H -1.000 *
6 F -1.000 *
7 V -1.000 *
8 P -1.000 *
9 Q -1.000 *
10 H -1.000 *
11 E -1.000 *
12 E -0.872
13 E -0.518
14 Q -0.837
15 V -0.112
16 Y 0.970
17 S 0.943
18 I 0.649
19 S -0.739
20 G 0.628
21 K 0.347
22 A 0.196
23 L -0.269
24 K 0.582
25 L 0.454
26 T 0.504
27 T 0.778
28 S -0.396
29 D -0.055
30 D 0.424
31 I 0.377
32 K -0.602
33 P -0.732
34 Y 0.132
35 L 0.548
36 E -0.535
37 E -0.804
38 L 1.197
39 A -0.938
40 A -0.659
41 L -1.315
42 K -0.897
43 T -1.188
44 C 0.493
45 T -0.557
46 K -0.921
47 L 1.827
48 D -0.535
49 L 1.526
50 S -0.404
51 G 1.555
52 N 1.704
53 T 1.525
54 I 1.145
55 G 2.301
56 T 0.035
57 E -0.840
58 A 1.484
59 S 1.264
60 E -0.821
61 A -0.747
62 L 1.057
63 A 1.388
64 K -0.745
65 C -0.475
66 I 1.520
67 A -0.690
68 E -0.675
69 N -0.639
70 T -0.917
71 Q -0.735
72 V -0.035
73 R 0.101
74 E -0.890
75 S -1.054
76 L 0.725
77 V -0.263
78 E -0.819
79 V 0.463
80 N -0.406
81 F 0.834
82 A -0.065
83 D 1.263
84 L 0.585
85 Y 1.408
86 T 0.568
87 S 0.985
88 R 2.885
89 L 0.006
90 V -0.328
91 D -0.315
92 E 2.427
93 V 1.158
94 V 1.477
95 D -1.041
96 S 1.058
97 L 1.695
98 K -0.771
99 F -0.678
100 L 1.245
101 L 0.125
102 P -0.592
103 V 0.293
104 L 0.839
105 L -0.854
106 K -0.677
107 C -0.735
108 P -0.734
109 H -0.706
110 L 1.701
111 E -0.783
112 I -0.237
113 V 1.971
114 N 0.478
115 L 1.520
116 S 0.629
117 D 1.062
118 N 3.889
119 A 1.530
120 F 1.546
121 G 2.257
122 L -0.684
123 R -0.659
124 T 0.200
125 I -0.252
126 E -0.944
127 L -0.356
128 L 1.171
129 E -0.641
130 D -0.820
131 Y 0.348
132 I 1.289
133 A -0.467
134 H -0.319
135 A -0.125
136 V -0.899
137 N -0.666
138 I 1.566
139 K 0.280
140 H -0.344
141 L 1.240
142 I -0.064
143 L 1.183
144 S -0.281
145 N 2.027
146 N 1.110
147 G 1.690
148 M 0.806
149 G 2.365
150 P -0.295
151 F -1.054
152 A 0.725
153 G 0.968
154 E -0.582
155 R -0.851
156 I 1.173
157 G 1.314
158 K -0.533
159 A 0.579
160 L 1.135
161 F -1.180
162 H -0.755
163 L 0.264
164 A -0.591
165 Q -1.033
166 N -0.730
167 K -0.717
168 K -0.841
169 A -0.689
170 A -0.827
171 S -0.439
172 K -1.069
173 P -0.691
174 F -1.136
175 L 1.716
176 E 0.154
177 T -0.322
178 F 1.301
179 I -0.610
180 C -0.034
181 G 1.437
182 R 0.963
183 N 1.196
184 R 0.942
185 L 1.041
186 E 1.161
187 N -0.046
188 G -0.541
189 S 1.230
190 A 0.438
191 V -1.152
192 Y -1.200
193 L 1.064
194 A 0.805
195 L -0.641
196 G -0.083
197 L 1.201
198 K -0.650
199 S -0.675
200 H -0.403
201 S -0.610
202 E -0.454
203 G -0.476
204 L 1.474
205 K 0.312
206 V -0.803
207 V 1.543
208 K -0.685
209 L 1.294
210 Y -0.826
211 Q 0.939
212 N 1.258
213 G 0.872
214 I 1.749
215 R -0.283
216 P -1.106
217 K -0.827
218 G 2.180
219 V -0.023
220 A -1.033
221 T -1.164
222 L 1.131
223 I 0.102
224 H -0.670
225 Y -1.050
226 G 0.758
227 L 0.442
228 Q -0.821
229 Y -1.128
230 L 0.109
231 K -0.696
232 N -0.814
233 L 2.370
234 E -0.491
235 I -0.511
236 L 1.466
237 D 0.919
238 L 1.239
239 Q 0.071
240 D 1.548
241 N 3.023
242 T -0.511
243 F -0.079
244 T -0.421
245 K -1.291
246 H -1.161
247 A 2.004
248 S -0.069
249 L -1.139
250 I -0.429
251 L 0.741
252 A 0.635
253 K -0.706
254 A -0.367
255 L 0.591
256 P -0.825
257 T -1.057
258 W -0.070
259 K -0.684
260 D -0.082
261 S -1.013
262 L 2.550
263 F -0.560
264 E -0.827
265 L 1.118
266 N 0.243
267 L 0.879
268 N 0.963
269 D 1.878
270 C 0.894
271 L -0.350
272 L 0.800
273 K -0.218
274 T -0.672
275 A -0.652
276 G 2.153
277 S -0.510
278 D -1.205
279 E -1.015
280 V 1.223
281 F -0.052
282 K -0.641
283 V -0.382
284 F 1.263
285 T -0.992
286 E -0.895
287 V -1.019
288 K -1.001
289 F -0.894
290 P -0.884
291 N -0.959
292 L 1.847
293 H -0.471
294 V -0.770
295 L 1.995
296 K -0.903
297 F 0.956
298 E -0.234
299 Y 0.248
300 N 1.321
301 E 0.577
302 M 1.294
303 A -0.697
304 Q -1.165
305 E -0.482
306 T 0.357
307 I -0.213
308 E -1.056
309 V -1.243
310 S -0.677
311 F -0.491
312 L -1.079
313 P -0.948
314 A -1.051
315 M -1.179
316 E -0.768
317 K -1.277
318 G -1.168
319 N -0.974
320 L -0.385
321 P -0.883
322 E -1.097
323 L 0.779
324 E -0.613
325 K -1.042
326 L 1.029
327 E -0.442
328 I 1.209
329 N 0.336
330 G -0.233
331 N 3.953
332 R -1.212
333 L 0.101
334 D -0.617
335 E -0.597
336 D -0.083
337 S -0.458
338 D -0.949
339 A -1.212
340 L -0.090
341 D -0.856
342 L -0.938
343 L 0.392
344 Q -0.886
345 S -0.710
346 K -1.098
347 F -0.646
348 D -0.664
349 D -0.863
350 L -1.103
351 E -0.636
352 V -0.986
353 D -0.602
354 D -0.999
355 F -0.860
356 E -0.841
357 E -0.834
358 V -0.895
359 D -0.706
360 S -0.868
361 E -0.056
362 D -0.323
363 E -0.902
364 E -0.722
365 G -0.384
366 E -0.830
367 D -0.437
368 E -0.546
369 E -0.683
370 D -0.565
371 E -0.578
372 D -0.503
373 E -0.487
374 D -0.329
375 E -0.455
376 K -0.921
377 L -0.890
378 E -0.744
379 E -0.889
380 I -1.016
381 E -0.706
382 T -0.795
383 E -0.439
384 R -0.808
385 L -1.018
386 E -0.723
387 K -0.777
388 E -0.677
389 L -1.000 *
390 L -0.826
391 E -0.218
392 V -0.809
393 Q -0.626
394 V 0.193
395 D 1.307
396 D -0.041
397 L 1.131
398 A 0.535
399 E -0.036
400 R -1.000 *
401 L -1.000 *
402 A -1.000 *
403 E -1.000 *
404 T -1.000 *
405 E -1.000 *
406 I -1.000 *
407 K -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 337.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MATLHFVPQH EEEQVYSISG KALKLTTSDD IKPYLEELAA LKTCTKLDLS GNTIGTEASE
70 80 90 100 110 120
ALAKCIAENT QVRESLVEVN FADLYTSRLV DEVVDSLKFL LPVLLKCPHL EIVNLSDNAF
130 140 150 160 170 180
GLRTIELLED YIAHAVNIKH LILSNNGMGP FAGERIGKAL FHLAQNKKAA SKPFLETFIC
190 200 210 220 230 240
GRNRLENGSA VYLALGLKSH SEGLKVVKLY QNGIRPKGVA TLIHYGLQYL KNLEILDLQD
250 260 270 280 290 300
NTFTKHASLI LAKALPTWKD SLFELNLNDC LLKTAGSDEV FKVFTEVKFP NLHVLKFEYN
310 320 330 340 350 360
EMAQETIEVS FLPAMEKGNL PELEKLEING NRLDEDSDAL DLLQSKFDDL EVDDFEEVDS
370 380 390 400
EDEEGEDEED EDEDEKLEEI ETERLEKELL EVQVDDLAER LAETEIK
Ran1p is yeast RanGAP1, the GTPase-activating protein for Ran which helps maintain the RanGTP/GDP gradient required for nucleocytoplasmic transport. Mouse homolog is listed in entry #338. In yeast, Ran1p is predominantly cytosolic. Two putative NES can be found in 316-357 and this region can direct reporter protein to the cytosol. The first NES motif 316-333 has less activity than the whole 316-357 fragment and mutation of L326 and L328 is sufficient to abolish export activity. The second NES motif 329-357 appears to have stronger NES activity than the former motif. CRM1 dependence was demonstrated by temperature sensitive CRM1 strain.
[1]. "Antagonistic Effects of NES and NLS Motifs Determine S. Cerevisiae Rna1p Subcellular Distribution"
Feng W, Benko AL, Lee JH, Stanford DR, Hopper AK. (1999)
J Cell Sci,
112(Pt3):339-47
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
