Ran GTPase-activating protein 1
UniProt
Show FASTA Format
>sp|P46061|RAGP1_MOUSE Ran GTPase-activating protein 1 OS=Mus musculus OX=10090 GN=Rangap1 PE=1 SV=2
MASEDIAKLAETLAKTQVAGGQLSFKGKGLKLNTAEDAKDVIKEIEEFDGLEALRLEGNT
VGVEAARVIAKALEKKSELKRCHWSDMFTGRLRSEIPPALISLGEGLITAGAQLVELDLS
DNAFGPDGVRGFEALLKSPACFTLQELKLNNCGMGIGGGKILAAALTECHRKSSAQGKPL
ALKVFVAGRNRLENDGATALAEAFGIIGTLEEVHMPQNGINHPGVTALAQAFAINPLLRV
INLNDNTFTEKGGVAMAETLKTLRQVEVINFGDCLVRSKGAVAIADAVRGGLPKLKELNL
SFCEIKRDAALVVAEAVADKAELEKLDLNGNALGEEGCEQLQEVMDSFNMAKVLASLSDD
EGEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGEEPATPSRKILDPN
SGEPAPVLSSPTPTDLSTFLSFPSPEKLLRLGPKVSVLIVQQTDTSDPEKVVSAFLKVAS
VFRDDASVKTAVLDAIDALMKKAFSCSSFNSNTFLTRLLIHMGLLKSEDKIKAIPSLHGP
LMVLNHVVRQDYFPKALAPLLLAFVTKPNGALETCSFARHNLLQTLYNI
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 972469999999974124784476715665889987499999996409993199807963
Pred: CCCHHHHHHHHHHHHHCCCCCEEEECCCCCCCCCHHHHHHHHHHHHCCCCCEEEEECCCC
AA: MASEDIAKLAETLAKTQVAGGQLSFKGKGLKLNTAEDAKDVIKEIEEFDGLEALRLEGNT
10 20 30 40 50 60
Conf: 439999999999812999525404332226674532899999999996189970088099
Pred: CCHHHHHHHHHHHHCCCCCCEEECCCCCCCCCCCCHHHHHHHHHHHHHHCCCCCEEECCC
AA: VGVEAARVIAKALEKKSELKRCHWSDMFTGRLRSEIPPALISLGEGLITAGAQLVELDLS
70 80 90 100 110 120
Conf: 998895882788984239998742011206899992379999999999999864079998
Pred: CCCCCCCCHHHHHHHHCCCCCCCCCEEECCCCCCCCHHHHHHHHHHHHHHHHHHHCCCCC
AA: DNAFGPDGVRGFEALLKSPACFTLQELKLNNCGMGIGGGKILAAALTECHRKSSAQGKPL
130 140 150 160 170 180
Conf: 851897188888870389999999852993599676899995879999999852999959
Pred: CCCEEECCCCCCCCCCHHHHHHHHHHCCCEEEEECCCCCCCHHHHHHHHHHHHHCCCCCE
AA: ALKVFVAGRNRLENDGATALAEAFGIIGTLEEVHMPQNGINHPGVTALAQAFAINPLLRV
190 200 210 220 230 240
Conf: 818898689326999999706799840884575345763099999998359999758986
Pred: EECCCCCCCCHHHHHHHHHHCCCCCCCEEECCCCCCCCCCHHHHHHHHHCCCCCCCEEEC
AA: INLNDNTFTEKGGVAMAETLKTLRQVEVINFGDCLVRSKGAVAIADAVRGGLPKLKELNL
250 260 270 280 290 300
Conf: 789889789999999960587776787669989943499999999726730001367766
Pred: CCCCCCHHHHHHHHHHHCCCCCCCEEECCCCCCCHHHHHHHHHHHHHCCCHHHCCCCCCC
AA: SFCEIKRDAALVVAEAVADKAELEKLDLNGNALGEEGCEQLQEVMDSFNMAKVLASLSDD
310 320 330 340 350 360
Conf: 799974112488776432333555664034443201479988899998899987557999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCC
AA: EGEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGEEPATPSRKILDPN
370 380 390 400 410 420
Conf: 999988889999699657520999888822597058999985189984799999984622
Pred: CCCCCCCCCCCCCCCHHHHHCCCCHHHHHCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHC
AA: SGEPAPVLSSPTPTDLSTFLSFPSPEKLLRLGPKVSVLIVQQTDTSDPEKVVSAFLKVAS
430 440 450 460 470 480
Conf: 115884578999999999999984345423236888999980644456777889997208
Pred: CCCCCHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCHH
AA: VFRDDASVKTAVLDAIDALMKKAFSCSSFNSNTFLTRLLIHMGLLKSEDKIKAIPSLHGP
490 500 510 520 530 540
Conf: 9999995615999723099999987187988765268779899988309
Pred: HHHHHHHHCCCCCCCCCHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHC
AA: LMVLNHVVRQDYFPKALAPLLLAFVTKPNGALETCSFARHNLLQTLYNI
550 560 570 580
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 S -1.000 *
4 E -0.498
5 D 0.513
6 I 0.297
7 A -0.709
8 K -0.422
9 L 1.659
10 A 0.156
11 E -0.012
12 T -0.067
13 L 1.520
14 A -1.000 *
15 K -0.424
16 T -0.314
17 Q -0.694
18 V -0.816
19 A -0.761
20 G -0.749
21 G -0.746
22 Q -0.807
23 L 1.560
24 S -0.170
25 F 0.516
26 K -1.157
27 G -0.216
28 K -0.131
29 G -0.357
30 L -0.028
31 K -0.060
32 L -1.077
33 N 0.352
34 T 0.512
35 A -0.423
36 E -0.480
37 D 0.176
38 A 0.598
39 K -0.821
40 D -0.748
41 V 0.497
42 I 0.352
43 K -0.507
44 E -0.527
45 I 2.134
46 E -0.752
47 E -0.918
48 F -1.009
49 D -0.917
50 G -0.881
51 L 1.127
52 E -0.410
53 A -1.070
54 L 1.892
55 R -0.977
56 L 2.087
57 E -0.896
58 G -0.483
59 N 0.926
60 T -0.584
61 V 1.398
62 G 1.360
63 V -0.314
64 E -0.810
65 A 1.419
66 A 0.823
67 R -0.610
68 V -0.087
69 I 1.863
70 A 1.114
71 K -0.386
72 A 0.071
73 L 2.263
74 E -0.132
75 K -0.936
76 K 0.012
77 S -0.767
78 E -0.892
79 L 1.858
80 K -0.396
81 R -1.020
82 C 1.299
83 H -0.892
84 W 1.105
85 S -0.836
86 D -0.677
87 M 0.296
88 F -0.975
89 T 0.777
90 G 0.208
91 R -0.355
92 L -0.885
93 R 0.108
94 S -0.053
95 E -0.133
96 I 0.865
97 P -0.201
98 P -0.985
99 A -0.155
100 L -0.089
101 I -0.972
102 S -0.741
103 L 1.254
104 G -0.187
105 E -0.549
106 G 0.982
107 L 1.046
108 I -0.621
109 T -0.688
110 A -0.676
111 G -0.432
112 A -0.328
113 Q -0.718
114 L 1.460
115 V -0.802
116 E -0.808
117 L 1.512
118 D -0.156
119 L 0.848
120 S -0.650
121 D -0.688
122 N 1.233
123 A -0.548
124 F 1.249
125 G 0.915
126 P -0.607
127 D -0.764
128 G -0.214
129 V -0.257
130 R -0.744
131 G -0.237
132 F 0.791
133 E -0.677
134 A -0.635
135 L 0.248
136 L 1.840
137 K -0.383
138 S -0.611
139 P -0.305
140 A -0.310
141 C -0.564
142 F -0.439
143 T 0.456
144 L 2.490
145 Q -0.562
146 E -0.866
147 L 1.806
148 K -0.890
149 L 1.269
150 N -0.994
151 N -0.578
152 C 1.449
153 G -0.565
154 M 1.008
155 G 0.672
156 I -0.689
157 G -1.041
158 G 1.481
159 G 0.827
160 K -0.817
161 I -0.188
162 L 1.411
163 A 1.137
164 A -0.499
165 A 1.007
166 L 2.373
167 T -0.893
168 E -0.734
169 C 0.351
170 H -0.769
171 R -1.102
172 K -0.611
173 S -0.308
174 S -0.695
175 A -0.488
176 Q -0.844
177 G 0.239
178 K -0.921
179 P -0.733
180 L -0.389
181 A -0.694
182 L 2.399
183 K -0.123
184 V -0.824
185 F 1.579
186 V -0.864
187 A 0.304
188 G -0.825
189 R -0.816
190 N 1.827
191 R -0.535
192 L 0.954
193 E 0.515
194 N -0.243
195 D -0.607
196 G 1.097
197 A 0.538
198 T -1.088
199 A -0.187
200 L 1.468
201 A 0.497
202 E -0.432
203 A 0.555
204 F 1.836
205 G -0.270
206 I -0.879
207 I 0.083
208 G -0.461
209 T -0.078
210 L 2.034
211 E -0.064
212 E -0.828
213 V 1.879
214 H -1.067
215 M 1.658
216 P -1.367
217 Q -0.830
218 N 1.451
219 G -0.423
220 I 2.017
221 N -0.587
222 H -1.205
223 P -0.879
224 G 2.757
225 V 0.330
226 T -0.865
227 A -0.453
228 L 1.897
229 A 0.189
230 Q -0.493
231 A 0.766
232 F 1.289
233 A -0.602
234 I -1.078
235 N 0.741
236 P -0.677
237 L -0.873
238 L 2.454
239 R -0.159
240 V -0.976
241 I 1.989
242 N -0.301
243 L 1.628
244 N -0.985
245 D -0.893
246 N 1.842
247 T -0.867
248 F 0.560
249 T -0.053
250 E -1.147
251 K -0.991
252 G 2.828
253 G 0.701
254 V -1.051
255 A -0.249
256 M 1.421
257 A 0.916
258 E -0.590
259 T 0.495
260 L 1.332
261 K -0.716
262 T -1.177
263 L 0.165
264 R -0.773
265 Q -1.026
266 V 2.493
267 E -0.245
268 V -0.984
269 I 1.876
270 N -0.321
271 F 1.254
272 G -0.834
273 D -0.693
274 C 1.374
275 L -0.577
276 V 1.260
277 R 0.304
278 S -0.632
279 K -0.672
280 G 2.697
281 A 0.682
282 V -1.151
283 A -0.697
284 I 1.528
285 A 0.214
286 D -0.477
287 A 0.074
288 V 1.329
289 R -0.681
290 G -1.077
291 G -0.394
292 L -0.461
293 P -0.859
294 K -1.119
295 L 2.078
296 K -0.435
297 E -0.895
298 L 1.901
299 N -0.983
300 L 1.609
301 S -0.924
302 F -0.944
303 C 1.314
304 E -0.689
305 I 1.463
306 K -0.531
307 R -1.225
308 D -0.826
309 A 0.990
310 A 0.296
311 L -1.041
312 V -0.891
313 V 1.665
314 A 0.202
315 E -0.715
316 A 0.093
317 V 0.103
318 A -0.829
319 D -0.773
320 K 0.071
321 A -1.042
322 E -1.014
323 L 1.557
324 E -0.675
325 K -1.075
326 L 1.566
327 D -0.675
328 L 2.083
329 N -0.894
330 G -0.762
331 N 2.681
332 A -1.323
333 L 1.113
334 G 0.359
335 E -0.904
336 E -0.430
337 G 0.386
338 C -0.274
339 E -0.842
340 Q -0.829
341 L 0.862
342 Q -0.780
343 E -0.335
344 V -0.613
345 M 0.351
346 D -0.656
347 S -0.975
348 F -0.942
349 N -0.905
350 M -1.094
351 A -0.626
352 K -0.741
353 V -1.064
354 L 0.683
355 A -1.077
356 S -0.411
357 L -1.034
358 S -0.825
359 D -0.325
360 D -0.916
361 E -0.998
362 G 0.160
363 E -0.571
364 D -0.662
365 E -1.000 *
366 D -1.000 *
367 E -1.000 *
368 E -1.000 *
369 E -1.000 *
370 E -1.000 *
371 G -1.000 *
372 E -1.000 *
373 E -1.000 *
374 D -1.000 *
375 D -1.000 *
376 E -1.000 *
377 E -1.000 *
378 E -1.000 *
379 E -1.000 *
380 D -1.000 *
381 E -1.000 *
382 E -1.000 *
383 D -1.000 *
384 E -1.000 *
385 E -1.000 *
386 D -1.000 *
387 D -1.000 *
388 D -1.000 *
389 E -1.000 *
390 E -1.000 *
391 E -1.000 *
392 E -1.000 *
393 E -1.000 *
394 Q -1.000 *
395 E -1.000 *
396 E -1.000 *
397 E -1.000 *
398 E -1.000 *
399 E -1.000 *
400 P -1.000 *
401 P -1.000 *
402 Q -1.000 *
403 R -1.000 *
404 G -1.000 *
405 S -1.000 *
406 G -1.000 *
407 E -1.000 *
408 E -1.000 *
409 P -1.000 *
410 A -1.000 *
411 T -1.000 *
412 P -1.000 *
413 S -1.000 *
414 R -1.000 *
415 K -1.000 *
416 I -1.000 *
417 L -1.000 *
418 D -1.000 *
419 P -1.000 *
420 N -1.000 *
421 S -1.000 *
422 G -1.000 *
423 E -1.000 *
424 P -1.000 *
425 A -1.000 *
426 P -1.000 *
427 V -1.000 *
428 L -1.000 *
429 S -1.000 *
430 S -1.000 *
431 P -1.000 *
432 T -1.000 *
433 P -1.000 *
434 T -1.000 *
435 D -1.000 *
436 L -1.000 *
437 S -1.000 *
438 T -1.000 *
439 F -1.000 *
440 L -1.000 *
441 S -1.000 *
442 F -1.000 *
443 P -1.000 *
444 S -1.000 *
445 P -1.000 *
446 E -1.000 *
447 K -1.000 *
448 L -1.000 *
449 L -1.000 *
450 R -1.000 *
451 L -1.000 *
452 G -1.000 *
453 P -1.000 *
454 K -1.000 *
455 V -1.000 *
456 S -1.000 *
457 V -1.000 *
458 L -1.000 *
459 I -1.000 *
460 V -1.000 *
461 Q -1.000 *
462 Q -1.000 *
463 T -1.000 *
464 D -1.000 *
465 T -1.000 *
466 S -1.000 *
467 D -1.000 *
468 P -1.000 *
469 E -1.000 *
470 K -1.000 *
471 V -1.000 *
472 V -1.000 *
473 S -1.000 *
474 A -1.000 *
475 F -1.000 *
476 L -1.000 *
477 K -1.000 *
478 V -1.000 *
479 A -1.000 *
480 S -1.000 *
481 V -1.000 *
482 F -1.000 *
483 R -1.000 *
484 D -1.000 *
485 D -1.000 *
486 A -1.000 *
487 S -1.000 *
488 V -1.000 *
489 K -1.000 *
490 T -1.000 *
491 A -1.000 *
492 V -1.000 *
493 L -1.000 *
494 D -1.000 *
495 A -1.000 *
496 I -1.000 *
497 D -1.000 *
498 A -1.000 *
499 L -1.000 *
500 M -1.000 *
501 K -1.000 *
502 K -1.000 *
503 A -1.000 *
504 F -1.000 *
505 S -1.000 *
506 C -1.000 *
507 S -1.000 *
508 S -1.000 *
509 F -1.000 *
510 N -1.000 *
511 S -1.000 *
512 N -1.000 *
513 T -1.000 *
514 F -1.000 *
515 L -1.000 *
516 T -1.000 *
517 R -1.000 *
518 L -1.000 *
519 L -1.000 *
520 I -1.000 *
521 H -1.000 *
522 M -1.000 *
523 G -1.000 *
524 L -1.000 *
525 L -1.000 *
526 K -1.000 *
527 S -1.000 *
528 E -1.000 *
529 D -1.000 *
530 K -1.000 *
531 I -1.000 *
532 K -1.000 *
533 A -1.000 *
534 I -1.000 *
535 P -1.000 *
536 S -1.000 *
537 L -1.000 *
538 H -1.000 *
539 G -1.000 *
540 P -1.000 *
541 L -1.000 *
542 M -1.000 *
543 V -1.000 *
544 L -1.000 *
545 N -1.000 *
546 H -1.000 *
547 V -1.000 *
548 V -1.000 *
549 R -1.000 *
550 Q -1.000 *
551 D -1.000 *
552 Y -1.000 *
553 F -1.000 *
554 P -1.000 *
555 K -1.000 *
556 A -1.000 *
557 L -1.000 *
558 A -1.000 *
559 P -1.000 *
560 L -1.000 *
561 L -1.000 *
562 L -1.000 *
563 A -1.000 *
564 F -1.000 *
565 V -1.000 *
566 T -1.000 *
567 K -1.000 *
568 P -1.000 *
569 N -1.000 *
570 G -1.000 *
571 A -1.000 *
572 L -1.000 *
573 E -1.000 *
574 T -1.000 *
575 C -1.000 *
576 S -1.000 *
577 F -1.000 *
578 A -1.000 *
579 R -1.000 *
580 H -1.000 *
581 N -1.000 *
582 L -1.000 *
583 L -1.000 *
584 Q -1.000 *
585 T -1.000 *
586 L -1.000 *
587 Y -1.000 *
588 N -1.000 *
589 I -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 338.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MASEDIAKLA ETLAKTQVAG GQLSFKGKGL KLNTAEDAKD VIKEIEEFDG LEALRLEGNT
70 80 90 100 110 120
VGVEAARVIA KALEKKSELK RCHWSDMFTG RLRSEIPPAL ISLGEGLITA GAQLVELDLS
130 140 150 160 170 180
DNAFGPDGVR GFEALLKSPA CFTLQELKLN NCGMGIGGGK ILAAALTECH RKSSAQGKPL
190 200 210 220 230 240
ALKVFVAGRN RLENDGATAL AEAFGIIGTL EEVHMPQNGI NHPGVTALAQ AFAINPLLRV
250 260 270 280 290 300
INLNDNTFTE KGGVAMAETL KTLRQVEVIN FGDCLVRSKG AVAIADAVRG GLPKLKELNL
310 320 330 340 350 360
SFCEIKRDAA LVVAEAVADK AELEKLDLNG NALGEEGCEQ LQEVMDSFNM AKVLASLSDD
370 380 390 400 410 420
EGEDEDEEEE GEEDDEEEED EEDEEDDDEE EEEQEEEEEP PQRGSGEEPA TPSRKILDPN
430 440 450 460 470 480
SGEPAPVLSS PTPTDLSTFL SFPSPEKLLR LGPKVSVLIV QQTDTSDPEK VVSAFLKVAS
490 500 510 520 530 540
VFRDDASVKT AVLDAIDALM KKAFSCSSFN SNTFLTRLLI HMGLLKSEDK IKAIPSLHGP
550 560 570 580
LMVLNHVVRQ DYFPKALAPL LLAFVTKPNG ALETCSFARH NLLQTLYNI
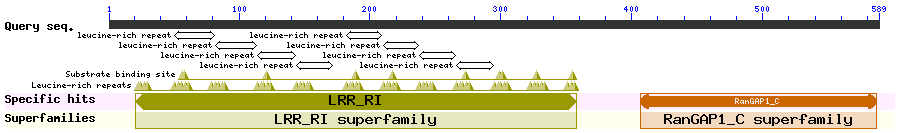
RanGAP1 is the GTPase-activating protein for Ran which helps maintain the RanGTP/GDP gradient required for nucleocytoplasmic transport. Matunis et al. found that RanGAP1 associates with the nuclear pore complex (NPC) for its function (requires a SUMO-1 modification) but can also be found in the cytosol. Its N-terminal region contains 9 leucine-rich repeats that resemble NESs (46-59, 109-122, 177-190, 205-218, 233-246, 261-274, 290-303, 318-331). This N-terminal region is conserved across different organisms. Yeast RanGAP1 is listed in entry #337, with region 316-357 shown to be functional NES. Cha et al. later found that RanGAP1 accumulates in the nucleus upon LMB treatment and CRM1 knockdown. LMB treatment also caused dissociation of the RanGAP1 from NPC to the nucleoplasm and an increase in RanGAP1 sumoylation.
Ref.1SUMO-1 Modification and Its Role in Targeting the Ran GTPase-activating Protein, RanGAP1, to the Nuclear Pore Complex, Martunis et al., J Cell Biol, 1998
[1]. "SUMO-1 Modification and Its Role in Targeting the Ran GTPase-activating Protein, RanGAP1, to the Nuclear Pore Complex"
Matunis MJ, Wu J, Blobel G. (1998)
J Cell Biol,
140(3):499-509
PubMed[2]. "The Cellular Distribution of RanGAP1 Is Regulated by CRM1-Mediated Nuclear Export in Mammalian Cells"
Cha K, Sen P, Raghunayakula S, Zhang XD. (2015)
PLoS One,
10(10):e0141309
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
