Carbonate dehydratase IX (CA-IX), Carbonic anhydrase IX (CAIX), Membrane antigen MN, P54/58N, Renal cell carcinoma-associated antigen G250, pMW1
Show FASTA Format
>sp|Q16790|CAH9_HUMAN Carbonic anhydrase 9 OS=Homo sapiens OX=9606 GN=CA9 PE=1 SV=2
MAPLCPSPWLPLLIPAPAPGLTVQLLLSLLLLVPVHPQRLPRMQEDSPLGGGSSGEDDPL
GEEDLPSEEDSPREEDPPGEEDLPGEEDLPGEEDLPEVKPKSEEEGSLKLEDLPTVEAPG
DPQEPQNNAHRDKEGDDQSHWRYGGDPPWPRVSPACAGRFQSPVDIRPQLAAFCPALRPL
ELLGFQLPPLPELRLRNNGHSVQLTLPPGLEMALGPGREYRALQLHLHWGAAGRPGSEHT
VEGHRFPAEIHVVHLSTAFARVDEALGRPGGLAVLAAFLEEGPEENSAYEQLLSRLEEIA
EEGSETQVPGLDISALLPSDFSRYFQYEGSLTTPPCAQGVIWTVFNQTVMLSAKQLHTLS
DTLWGPGDSRLQLNFRATQPLNGRVIEASFPAGVDSSPRAAEPVQLNSCLAAGDILALVF
GLLFAVTSVAFLVQMRRQHRRGTKGGVSYRPAEVAETGA
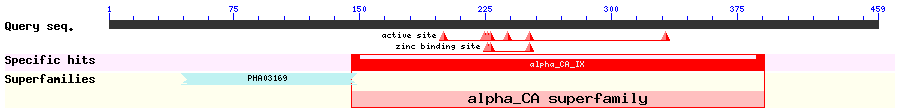
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 999999999876236899981699999998634589999777757899999999999999
Pred: CCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: MAPLCPSPWLPLLIPAPAPGLTVQLLLSLLLLVPVHPQRLPRMQEDSPLGGGSSGEDDPL
10 20 30 40 50 60
Conf: 999999998999999999999999999999999999999997433454447898767999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: GEEDLPSEEDSPREEDPPGEEDLPGEEDLPGEEDLPEVKPKSEEEGSLKLEDLPTVEAPG
70 80 90 100 110 120
Conf: 999999887778999999885589999999989989898889765447950108999872
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCEEECCCCCCC
AA: DPQEPQNNAHRDKEGDDQSHWRYGGDPPWPRVSPACAGRFQSPVDIRPQLAAFCPALRPL
130 140 150 160 170 180
Conf: 126879999998011028825875179876333489972012133210389999997554
Pred: CCCCCCCCCCCCCEEECCCEEEEEECCCCCCCCCCCCCCCHHHHHHHCCCCCCCCCCCCC
AA: ELLGFQLPPLPELRLRNNGHSVQLTLPPGLEMALGPGREYRALQLHLHWGAAGRPGSEHT
190 200 210 220 230 240
Conf: 488435316769740688999155508999357653302309998924899998875301
Pred: CCCCCCCEEEEEEECCCCCCCHHHHHCCCCCEEEEEEEHHCCCCCCCHHHHHHHHHHHCC
AA: VEGHRFPAEIHVVHLSTAFARVDEALGRPGGLAVLAAFLEEGPEENSAYEQLLSRLEEIA
250 260 270 280 290 300
Conf: 499243105875454799897984000366789998786027882530103599999999
Pred: CCCCEEECCCCCCCCCCCCCCCCCEECCCCCCCCCCCCCEEEEEECCHHHCCHHHHHHHH
AA: EEGSETQVPGLDISALLPSDFSRYFQYEGSLTTPPCAQGVIWTVFNQTVMLSAKQLHTLS
310 320 330 340 350 360
Conf: 812999997000267889899997899702599899998889743454332101456534
Pred: HHCCCCCCCCCCCCCCCCCCCCCCEEEEECCCCCCCCCCCCCCCCCCCCCCCHHHHHHHH
AA: DTLWGPGDSRLQLNFRATQPLNGRVIEASFPAGVDSSPRAAEPVQLNSCLAAGDILALVF
370 380 390 400 410 420
Conf: 454433567888888765259999972762642322589
Pred: HHHHHHHHHHHHHHHHHHCCCCCCCCEEEECCCCCCCCC
AA: GLLFAVTSVAFLVQMRRQHRRGTKGGVSYRPAEVAETGA
430 440 450
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 P -1.000 *
4 L -1.000 *
5 C -1.000 *
6 P -1.000 *
7 S -1.000 *
8 P -1.000 *
9 W -1.000 *
10 L -1.000 *
11 P -1.000 *
12 L -1.000 *
13 L -1.000 *
14 I -1.000 *
15 P -1.000 *
16 A -1.000 *
17 P -1.000 *
18 A -1.000 *
19 P -1.000 *
20 G -1.000 *
21 L -1.000 *
22 T -1.000 *
23 V -1.000 *
24 Q -1.000 *
25 L -1.000 *
26 L -1.000 *
27 L -1.000 *
28 S -1.000 *
29 L -1.000 *
30 L -1.000 *
31 L -1.000 *
32 L -1.000 *
33 V -1.000 *
34 P -1.000 *
35 V -1.000 *
36 H -1.000 *
37 P -1.000 *
38 Q -1.000 *
39 R -1.000 *
40 L -1.000 *
41 P -1.000 *
42 R -1.000 *
43 M -1.000 *
44 Q -1.000 *
45 E -1.000 *
46 D -1.000 *
47 S -1.000 *
48 P -1.000 *
49 L -1.000 *
50 G -1.000 *
51 G -1.000 *
52 G -1.000 *
53 S -1.000 *
54 S -1.000 *
55 G -1.000 *
56 E -1.000 *
57 D -1.000 *
58 D -1.000 *
59 P -1.000 *
60 L -1.000 *
61 G -1.000 *
62 E -1.000 *
63 E -1.000 *
64 D -1.000 *
65 L -1.000 *
66 P -1.000 *
67 S -1.000 *
68 E -1.000 *
69 E -1.000 *
70 D -1.000 *
71 S -1.000 *
72 P -1.000 *
73 R -1.000 *
74 E -1.000 *
75 E -1.000 *
76 D -1.000 *
77 P -1.000 *
78 P -1.000 *
79 G -1.000 *
80 E -1.000 *
81 E -1.000 *
82 D -1.000 *
83 L -1.000 *
84 P -1.000 *
85 G -1.000 *
86 E -1.000 *
87 E -1.000 *
88 D -1.000 *
89 L -1.000 *
90 P -1.000 *
91 G -1.000 *
92 E -1.000 *
93 E -1.000 *
94 D -1.000 *
95 L -1.000 *
96 P -1.000 *
97 E -1.000 *
98 V -1.000 *
99 K -1.000 *
100 P -1.000 *
101 K -1.000 *
102 S -1.000 *
103 E -1.000 *
104 E -1.000 *
105 E -1.000 *
106 G -1.000 *
107 S -1.000 *
108 L -1.000 *
109 K -1.000 *
110 L -1.000 *
111 E -1.000 *
112 D -1.000 *
113 L -1.000 *
114 P -1.000 *
115 T -1.000 *
116 V -1.000 *
117 E -1.000 *
118 A -1.000 *
119 P -1.000 *
120 G -1.000 *
121 D -1.000 *
122 P -1.000 *
123 Q -1.000 *
124 E -1.000 *
125 P -1.000 *
126 Q -1.000 *
127 N -1.000 *
128 N -1.000 *
129 A -1.000 *
130 H -1.000 *
131 R -1.000 *
132 D -1.000 *
133 K -1.000 *
134 E -1.000 *
135 G -1.000 *
136 D -1.000 *
137 D -0.571
138 Q -0.334
139 S -0.565
140 H -0.635
141 W -0.468
142 R -0.083
143 Y -0.114
144 G -0.847
145 G -0.589
146 D -0.646
147 P -0.907
148 P -0.912
149 W 1.934
150 P -0.973
151 R -0.828
152 V -1.128
153 S -0.404
154 P -0.479
155 A -0.909
156 C 1.431
157 A -0.530
158 G 0.075
159 R -0.997
160 F -0.718
161 Q 2.476
162 S 2.476
163 P 2.476
164 V 1.391
165 D 0.460
166 I 0.903
167 R -0.958
168 P -0.799
169 Q -0.805
170 L -0.987
171 A 0.005
172 A -0.927
173 F -0.334
174 C -0.121
175 P -0.858
176 A -0.781
177 L 1.252
178 R -0.935
179 P -0.582
180 L 0.339
181 E -1.034
182 L 0.044
183 L -1.134
184 G 1.016
185 F 1.254
186 Q -0.506
187 L -0.805
188 P -0.882
189 P -1.042
190 L -1.027
191 P -0.952
192 E -1.010
193 L -0.650
194 R -0.968
195 L 0.694
196 R -0.998
197 N 2.178
198 N 0.735
199 G 2.167
200 H 1.008
201 S 0.663
202 V 0.080
203 Q -0.621
204 L 0.585
205 T -0.895
206 L 0.513
207 P -0.731
208 P -0.684
209 G -0.910
210 L -0.945
211 E -1.205
212 M -0.708
213 A -0.869
214 L -0.834
215 G -0.315
216 P -1.000
217 G -0.510
218 R -0.908
219 E -0.966
220 Y 1.198
221 R -0.439
222 A 0.728
223 L -0.835
224 Q 1.700
225 L 0.539
226 H 2.152
227 L 0.521
228 H 2.152
229 W 2.158
230 G 2.098
231 A -0.719
232 A -1.074
233 G -0.980
234 R -1.181
235 P -1.057
236 G 1.202
237 S 0.984
238 E 1.063
239 H 1.107
240 T -0.621
241 V 0.256
242 E -0.128
243 G -0.187
244 H -0.952
245 R -0.708
246 F -0.143
247 P -0.510
248 A -0.317
249 E 1.282
250 I 0.175
251 H 1.567
252 V 0.374
253 V 0.865
254 H 0.749
255 L -0.143
256 S 0.380
257 T -0.367
258 A -0.465
259 F 0.333
260 A -0.743
261 R -0.144
262 V -0.512
263 D -0.885
264 E -0.765
265 A 1.131
266 L -0.464
267 G -1.017
268 R -0.587
269 P -0.584
270 G -0.481
271 G 1.304
272 L 0.668
273 A 0.511
274 V 1.049
275 L 0.457
276 A 1.096
277 A 0.153
278 F 0.964
279 L -0.222
280 E -0.338
281 E -0.462
282 G -0.573
283 P -0.881
284 E -0.963
285 E -0.876
286 N -0.348
287 S -0.763
288 A -0.870
289 Y 0.476
290 E -0.541
291 Q -0.835
292 L 0.742
293 L 0.185
294 S -0.387
295 R -0.840
296 L 0.579
297 E -0.966
298 E -0.893
299 I 1.255
300 A -0.592
301 E -0.922
302 E -0.555
303 G 0.463
304 S -0.736
305 E -0.916
306 T -0.379
307 Q -1.217
308 V 0.339
309 P -0.856
310 G -0.510
311 L 0.119
312 D -0.156
313 I 0.113
314 S -0.851
315 A -0.863
316 L 0.926
317 L 1.112
318 P 2.075
319 S -0.978
320 D -0.589
321 F -0.298
322 S -0.992
323 R -0.907
324 Y 1.426
325 F 0.941
326 Q 0.715
327 Y 2.476
328 E -0.976
329 G 2.476
330 S 2.476
331 L 1.546
332 T 2.156
333 T 1.671
334 P 2.476
335 P 0.352
336 C 1.682
337 A -0.450
338 Q 0.991
339 G -0.148
340 V 1.816
341 I -0.494
342 W 2.476
343 T 0.408
344 V 0.880
345 F 0.365
346 N -0.690
347 Q -0.399
348 T -0.430
349 V 0.355
350 M -1.021
351 L 0.510
352 S 0.566
353 A -1.129
354 K -0.783
355 Q 2.476
356 L 0.111
357 H -1.074
358 T -0.741
359 L 0.866
360 S -0.538
361 D -0.782
362 T -0.562
363 L -0.654
364 W -0.776
365 G -0.519
366 P -0.854
367 G -0.932
368 D -0.811
369 S -0.956
370 R -1.097
371 L 1.175
372 Q -0.823
373 L -0.628
374 N 0.496
375 F 0.388
376 R 2.205
377 A -0.981
378 T -0.286
379 Q 1.950
380 P -0.493
381 L 0.225
382 N -0.242
383 G -0.557
384 R 2.206
385 V -0.978
386 I 1.066
387 E -0.603
388 A -0.158
389 S 0.915
390 F 0.500
391 P -0.641
392 A -0.834
393 G -1.000 *
394 V -1.000 *
395 D -1.000 *
396 S -1.000 *
397 S -1.000 *
398 P -1.000 *
399 R -1.000 *
400 A -1.000 *
401 A -1.000 *
402 E -1.000 *
403 P -1.000 *
404 V -1.000 *
405 Q -1.000 *
406 L -1.000 *
407 N -1.000 *
408 S -1.000 *
409 C -1.000 *
410 L -1.000 *
411 A -1.000 *
412 A -1.000 *
413 G -1.000 *
414 D -1.000 *
415 I 0.056
416 L -0.110
417 A 0.316
418 L -0.214
419 V -0.034
420 F 0.141
421 G -1.000 *
422 L -1.000 *
423 L -1.000 *
424 F -1.000 *
425 A -1.000 *
426 V -1.000 *
427 T -1.000 *
428 S -1.000 *
429 V -1.000 *
430 A -1.000 *
431 F -1.000 *
432 L -1.000 *
433 V -1.000 *
434 Q -1.000 *
435 M -1.000 *
436 R -1.000 *
437 R -1.000 *
438 Q -1.000 *
439 H -1.000 *
440 R -1.000 *
441 R -1.000 *
442 G -1.000 *
443 T -1.000 *
444 K -1.000 *
445 G -1.000 *
446 G -1.000 *
447 V -1.000 *
448 S -1.000 *
449 Y -1.000 *
450 R -1.000 *
451 P -1.000 *
452 A -1.000 *
453 E -1.000 *
454 V -1.000 *
455 A -1.000 *
456 E -1.000 *
457 T -1.000 *
458 G -1.000 *
459 A -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 340.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MAPLCPSPWL PLLIPAPAPG LTVQLLLSLL LLVPVHPQRL PRMQEDSPLG GGSSGEDDPL
70 80 90 100 110 120
GEEDLPSEED SPREEDPPGE EDLPGEEDLP GEEDLPEVKP KSEEEGSLKL EDLPTVEAPG
130 140 150 160 170 180
DPQEPQNNAH RDKEGDDQSH WRYGGDPPWP RVSPACAGRF QSPVDIRPQL AAFCPALRPL
190 200 210 220 230 240
ELLGFQLPPL PELRLRNNGH SVQLTLPPGL EMALGPGREY RALQLHLHWG AAGRPGSEHT
250 260 270 280 290 300
VEGHRFPAEI HVVHLSTAFA RVDEALGRPG GLAVLAAFLE EGPEENSAYE QLLSRLEEIA
310 320 330 340 350 360
EEGSETQVPG LDISALLPSD FSRYFQYEGS LTTPPCAQGV IWTVFNQTVM LSAKQLHTLS
370 380 390 400 410 420
DTLWGPGDSR LQLNFRATQP LNGRVIEASF PAGVDSSPRA AEPVQLNSCL AAGDILALVF
430 440 450
GLLFAVTSVA FLVQMRRQHR RGTKGGVSYR PAEVAETGA
2HKF (X-Ray,2.01 Å resolution)
5DVX (X-Ray,1.6 Å resolution)
CA IX is a transmembrane protein at the cell surface with an extracellular catalytic domain, but have also been found in other compartments such as the nucleus. CRM1 was identified as an interactor of CA IX through IP experiments and the interaction was found to be dependent on the C-terminal region of CA IX. Under hypoxia, CRM1 and CA IX are found to redistribute to the nucleoli and co-localize in a LMB-sensitive manner. Putative NES identified in 412-429 can direct nuclear exit of EGFP fusion protein in LMB-sensitive manner. The sequence does not have a clear NES consensus match as there is an excess of hydrophobic residues.
[1]. "Binding of Carbonic Anhydrase IX to 45S rDNA Genes Is Prevented by exportin-1 in Hypoxic Cells"
Sasso E, Vitale M, Monteleone F, Boffo FL, Santoriello M, Sarnataro D, Garbi C, Sabatella M, Crifò B, Paolella LA, Minopoli G, Winum JY, Zambrano N. (2015)
Biomed Res Int,
2015:674920
PubMed[2]. "Characterization of Carbonic Anhydrase IX Interactome Reveals Proteins Assisting Its Nuclear Localization in Hypoxic Cells"
Buanne P, Renzone G, Monteleone F, Vitale M, Monti SM, Sandomenico A, Garbi C, Montanaro D, Accardo M, Troncone G, Zatovicova M, Csaderova L, Supuran CT, Pastorekova S, Scaloni A, De Simone G, Zambrano N. (2013)
J Proteome Res,
12(1):282-92
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
