Show FASTA Format
>sp|P19532|TFE3_HUMAN Transcription factor E3 OS=Homo sapiens OX=9606 GN=TFE3 PE=1 SV=4
MSHAAEPARDGVEASAEGPRAVFVLLEERRPADSAQLLSLNSLLPESGIVADIELENVLD
PDSFYELKSQPLPLRSSLPISLQATPATPATLSASSSAGGSRTPAMSSSSSSRVLLRQQL
MRAQAQEQERRERREQAAAAPFPSPAPASPAISVVGVSAGGHTLSRPPPAQVPREVLKVQ
THLENPTRYHLQQARRQQVKQYLSTTLGPKLASQALTPPPGPASAQPLPAPEAAHTTGPT
GSAPNSPMALLTIGSSSEKEIDDVIDEIISLESSYNDEMLSYLPGGTTGLQLPSTLPVSG
NLLDVYSSQGVATPAITVSNSCPAELPNIKREISETEAKALLKERQKKDNHNLIERRRRF
NINDRIKELGTLIPKSSDPEMRWNKGTILKASVDYIRKLQKEQQRSKDLESRQRSLEQAN
RSLQLRIQELELQAQIHGLPVPPTPGLLSLATTSASDSLKPEQLDIEEEGRPGAATFHVG
GGPAQNAPHQQPPAPPSDALLDLHFPSDHLGDLGDPFHLGLEDILMEEEEGVVGGLSGGA
LSPLRAASDPLLSSVSPAVSKASSRRSSFSMEEES
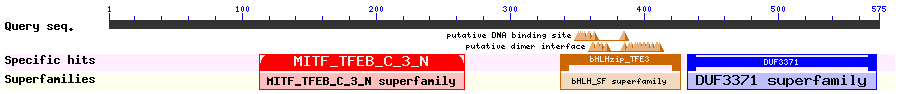
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 998887587887643479948999941589765324122445677863001202033249
Pred: CCCCCCCCCCCCCCCCCCCCEEEEEECCCCCCCCCCCCCCCCCCCCCCCCCCEEECCCCC
AA: MSHAAEPARDGVEASAEGPRAVFVLLEERRPADSAQLLSLNSLLPESGIVADIELENVLD
10 20 30 40 50 60
Conf: 875200046568899998844566889876555677899999999986315678999999
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHH
AA: PDSFYELKSQPLPLRSSLPISLQATPATPATLSASSSAGGSRTPAMSSSSSSRVLLRQQL
70 80 90 100 110 120
Conf: 999999999999999996099999999999713114589998899999998891155410
Pred: HHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCEECCCCCCCCCCCCCCCCCCCCEEEEEE
AA: MRAQAQEQERRERREQAAAAPFPSPAPASPAISVVGVSAGGHTLSRPPPAQVPREVLKVQ
130 140 150 160 170 180
Conf: 024596156898999988999863214852223445899999999999999777789999
Pred: CCCCCCHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: THLENPTRYHLQQARRQQVKQYLSTTLGPKLASQALTPPPGPASAQPLPAPEAAHTTGPT
190 200 210 220 230 240
Conf: 999998014410589961003557665550344332121114799997776788757777
Pred: CCCCCCCHHHHHCCCCCCHHHHHHHHHHHHCCCCCCHHHHCCCCCCCCCCCCCCCCCCCC
AA: GSAPNSPMALLTIGSSSEKEIDDVIDEIISLESSYNDEMLSYLPGGTTGLQLPSTLPVSG
250 260 270 280 290 300
Conf: 742002899988984010268999998631004499999999875103461144432037
Pred: CCCCCCCCCCCCCCCEECCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCCCCHHHHHHHCC
AA: NLLDVYSSQGVATPAITVSNSCPAELPNIKREISETEAKALLKERQKKDNHNLIERRRRF
310 320 330 340 350 360
Conf: 841234431146668999553346564421447999987665444344443223332012
Pred: CCCHHHHHHCCCCCCCCCCCCCCCCCCEEHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCC
AA: NINDRIKELGTLIPKSSDPEMRWNKGTILKASVDYIRKLQKEQQRSKDLESRQRSLEQAN
370 380 390 400 410 420
Conf: 102345467777888608999999999811100355568965346432479997544569
Pred: CHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: RSLQLRIQELELQAQIHGLPVPPTPGLLSLATTSASDSLKPEQLDIEEEGRPGAATFHVG
430 440 450 460 470 480
Conf: 999999999999999997754666799888999998888986411001113457888998
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: GGPAQNAPHQQPPAPPSDALLDLHFPSDHLGDLGDPFHLGLEDILMEEEEGVVGGLSGGA
490 500 510 520 530 540
Conf: 88988889998778895765657876775454589
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: LSPLRAASDPLLSSVSPAVSKASSRRSSFSMEEES
550 560 570
Show Conservation Score by AL2CO
1 M -1.000 *
2 S -1.000 *
3 H -1.000 *
4 A -1.000 *
5 A -1.000 *
6 E -1.000 *
7 P -1.000 *
8 A -1.000 *
9 R -1.000 *
10 D -1.000 *
11 G -1.000 *
12 V -1.000 *
13 E -1.000 *
14 A -1.000 *
15 S -1.000 *
16 A -1.000 *
17 E -1.000 *
18 G -1.000 *
19 P -1.000 *
20 R -1.000 *
21 A -1.000 *
22 V -1.000 *
23 F -1.000 *
24 V -1.000 *
25 L -1.000 *
26 L -1.000 *
27 E -1.000 *
28 E -1.000 *
29 R -1.000 *
30 R -1.000 *
31 P -1.000 *
32 A -1.000 *
33 D -1.000 *
34 S -1.000 *
35 A -1.000 *
36 Q -1.000 *
37 L -1.000 *
38 L -1.000 *
39 S -1.000 *
40 L -1.000 *
41 N -1.000 *
42 S -1.000 *
43 L -1.000 *
44 L -1.000 *
45 P -1.000 *
46 E -1.000 *
47 S -1.000 *
48 G -1.000 *
49 I -1.000 *
50 V -1.000 *
51 A -1.000 *
52 D -1.000 *
53 I -1.000 *
54 E -1.000 *
55 L -1.000 *
56 E -1.000 *
57 N -1.000 *
58 V -1.000 *
59 L -1.000 *
60 D -1.000 *
61 P -1.000 *
62 D -1.000 *
63 S -1.000 *
64 F -1.000 *
65 Y -1.000 *
66 E -1.000 *
67 L -1.000 *
68 K -1.000 *
69 S -1.000 *
70 Q -1.000 *
71 P -1.000 *
72 L -1.000 *
73 P -1.000 *
74 L -1.000 *
75 R -1.000 *
76 S -1.000 *
77 S -1.000 *
78 L -1.000 *
79 P -1.000 *
80 I -1.000 *
81 S -1.000 *
82 L -1.000 *
83 Q -1.000 *
84 A -1.000 *
85 T -1.000 *
86 P -1.000 *
87 A -1.000 *
88 T -1.000 *
89 P -1.000 *
90 A -1.000 *
91 T -1.000 *
92 L -1.000 *
93 S -1.000 *
94 A -1.000 *
95 S -1.000 *
96 S -1.000 *
97 S -1.000 *
98 A -1.000 *
99 G -1.000 *
100 G -1.000 *
101 S -1.000 *
102 R -1.000 *
103 T -1.000 *
104 P -1.000 *
105 A -1.000 *
106 M -1.000 *
107 S -1.000 *
108 S -1.000 *
109 S -1.000 *
110 S 0.006
111 S -0.101
112 S -0.111
113 R 0.516
114 V 0.328
115 L -0.279
116 L -0.294
117 R 0.261
118 Q 0.204
119 Q 0.136
120 L 1.049
121 M -0.049
122 R 1.003
123 A 0.394
124 Q -0.067
125 A -0.140
126 Q -0.260
127 E -0.068
128 Q 0.637
129 E 0.069
130 R -0.414
131 R -0.085
132 E -0.540
133 R -0.453
134 R -0.742
135 E -0.720
136 Q -0.540
137 A -0.534
138 A -0.714
139 A -0.753
140 A -0.672
141 P -0.695
142 F -0.853
143 P -0.749
144 S -0.786
145 P -0.820
146 A -0.814
147 P -0.655
148 A -0.570
149 S -0.580
150 P -0.182
151 A -0.588
152 I -0.487
153 S -0.654
154 V -0.383
155 V -0.561
156 G -0.661
157 V -0.386
158 S -0.695
159 A -0.639
160 G -0.672
161 G -0.501
162 H -1.000 *
163 T -1.000 *
164 L -0.592
165 S -0.095
166 R -0.609
167 P -0.375
168 P -0.679
169 P -0.464
170 A -0.532
171 Q -0.685
172 V 0.214
173 P -0.179
174 R -0.905
175 E -0.138
176 V -0.535
177 L -0.697
178 K -0.525
179 V 0.073
180 Q 0.102
181 T -0.046
182 H -0.595
183 L 0.410
184 E 0.453
185 N 0.417
186 P 0.740
187 T 0.215
188 R -0.428
189 Y -0.054
190 H 0.034
191 L 0.122
192 Q -0.394
193 Q 0.341
194 A 0.101
195 R 0.281
196 R -0.467
197 Q -0.248
198 Q 0.687
199 V 0.199
200 K -0.638
201 Q -0.616
202 Y -0.264
203 L -0.254
204 S -0.578
205 T -0.439
206 T -0.337
207 L -0.642
208 G -0.684
209 P -0.480
210 K -0.805
211 L -0.871
212 A -0.252
213 S -0.497
214 Q -0.612
215 A -0.544
216 L -0.781
217 T -0.676
218 P -0.710
219 P -0.550
220 P -0.591
221 G -0.543
222 P -0.608
223 A -0.653
224 S -0.628
225 A -0.806
226 Q -0.728
227 P -0.460
228 L -0.879
229 P -0.626
230 A -0.806
231 P -0.609
232 E -0.904
233 A -0.670
234 A -0.832
235 H -0.435
236 T -0.717
237 T -0.606
238 G -0.639
239 P -0.614
240 T -0.694
241 G -0.536
242 S -0.755
243 A -0.666
244 P -0.367
245 N -0.608
246 S -0.099
247 P -0.328
248 M -0.462
249 A -0.432
250 L -0.559
251 L -0.513
252 T -0.590
253 I -0.236
254 G -0.594
255 S -0.668
256 S -0.565
257 S -0.717
258 E -0.594
259 K -0.877
260 E -0.818
261 I -0.563
262 D -0.274
263 D -0.710
264 V -0.336
265 I -0.644
266 D 0.090
267 E -0.293
268 I -0.571
269 I -0.542
270 S -0.088
271 L -0.451
272 E -0.480
273 S 0.137
274 S -0.249
275 Y -0.800
276 N -0.494
277 D -0.703
278 E -0.842
279 M -0.489
280 L -0.823
281 S -0.339
282 Y -0.926
283 L -0.738
284 P -1.000 *
285 G -1.000 *
286 G -0.653
287 T -0.673
288 T -0.418
289 G -0.800
290 L -0.561
291 Q -0.749
292 L -0.520
293 P -0.507
294 S -0.608
295 T -0.354
296 L -0.668
297 P -0.036
298 V -0.591
299 S -0.270
300 G -0.503
301 N -0.403
302 L -0.234
303 L -0.627
304 D 0.454
305 V 0.142
306 Y -0.585
307 S -0.437
308 S -0.615
309 Q -0.508
310 G -0.464
311 V -0.616
312 A -0.364
313 T -0.438
314 P -0.506
315 A -0.347
316 I -0.658
317 T 0.034
318 V -0.087
319 S 0.139
320 N -0.209
321 S 0.077
322 C -0.627
323 P 0.158
324 A -0.190
325 E 0.311
326 L -0.028
327 P -0.529
328 N -0.581
329 I 0.076
330 K 0.006
331 R -0.652
332 E -0.395
333 I -0.131
334 S -0.562
335 E 0.027
336 T -0.119
337 E 0.131
338 A -0.301
339 K -0.671
340 A -0.065
341 L -0.362
342 L -0.218
343 K 0.351
344 E 0.797
345 R 1.809
346 Q 0.944
347 K 2.169
348 K 2.166
349 D 0.815
350 N 0.933
351 H 1.519
352 N 2.330
353 L 0.497
354 I 1.648
355 E 2.174
356 R 1.908
357 R 1.916
358 R 2.703
359 R 3.076
360 F 1.424
361 N 1.498
362 I 3.145
363 N 2.519
364 D 1.428
365 R 2.025
366 I 2.925
367 K 0.282
368 E 2.266
369 L 3.810
370 G 1.419
371 T -0.105
372 L 1.975
373 I 1.511
374 P 1.567
375 K 1.342
376 S 0.134
377 S 0.043
378 D 1.175
379 P 0.339
380 E 1.047
381 M 0.355
382 R 2.435
383 W 0.418
384 N 3.043
385 K 3.286
386 G 3.295
387 T 1.125
388 I 2.597
389 L 3.379
390 K 2.595
391 A 0.795
392 S 2.046
393 V 1.854
394 D 2.390
395 Y 2.260
396 I 2.565
397 R 1.719
398 K -0.471
399 L 2.717
400 Q 2.216
401 K 0.694
402 E 0.648
403 Q 0.161
404 Q -0.307
405 R 1.727
406 S 0.398
407 K 0.194
408 D 0.370
409 L -0.080
410 E 1.125
411 S -0.740
412 R 0.183
413 Q 0.398
414 R 1.544
415 S 0.182
416 L 1.021
417 E 0.680
418 Q -0.095
419 A -0.371
420 N 1.561
421 R 1.123
422 S -0.099
423 L 2.109
424 Q -0.134
425 L 0.213
426 R 0.944
427 I 1.279
428 Q 0.922
429 E 0.867
430 L 2.372
431 E 1.377
432 L -0.185
433 Q 0.522
434 A 0.780
435 Q 0.167
436 I -0.190
437 H 0.696
438 G 1.390
439 L 1.234
440 P 0.037
441 V -0.223
442 P -0.200
443 P -0.294
444 T -0.507
445 P -0.465
446 G -0.709
447 L -0.592
448 L -0.759
449 S -0.690
450 L -0.791
451 A -0.770
452 T -0.606
453 T -0.877
454 S -0.870
455 A -0.795
456 S -0.837
457 D -0.824
458 S -0.589
459 L -0.811
460 K -0.808
461 P -0.779
462 E -0.609
463 Q -0.855
464 L -0.889
465 D -0.755
466 I -0.883
467 E -0.846
468 E -0.816
469 E -0.996
470 G -0.820
471 R -0.870
472 P -0.787
473 G -0.956
474 A -0.919
475 A -0.878
476 T -0.919
477 F -0.934
478 H -0.975
479 V -0.889
480 G -0.866
481 G -0.901
482 G -0.972
483 P -0.852
484 A -0.928
485 Q -0.878
486 N -0.870
487 A -0.847
488 P -0.354
489 H -0.749
490 Q -0.847
491 Q -0.911
492 P -0.813
493 P -0.680
494 A -0.754
495 P -0.689
496 P -0.702
497 S -0.930
498 D -0.752
499 A -0.594
500 L -0.778
501 L -0.797
502 D -0.346
503 L -0.786
504 H -0.748
505 F -0.718
506 P -0.337
507 S -0.734
508 D -0.652
509 H -0.250
510 L -0.639
511 G -0.702
512 D -0.249
513 L -0.603
514 G -0.512
515 D -0.783
516 P -0.323
517 F -0.591
518 H -0.822
519 L -0.678
520 G -0.467
521 L -0.584
522 E -0.405
523 D -0.287
524 I -0.518
525 L -0.214
526 M -0.730
527 E 0.403
528 E 0.627
529 E -0.550
530 E -1.000 *
531 G -1.000 *
532 V -1.000 *
533 V -1.000 *
534 G -1.000 *
535 G -1.000 *
536 L -1.000 *
537 S -1.000 *
538 G -0.400
539 G -0.493
540 A -0.427
541 L -0.007
542 S 1.370
543 P 1.447
544 L 0.189
545 R -1.000 *
546 A -1.000 *
547 A 0.678
548 S -0.116
549 D 2.962
550 P 1.931
551 L 1.982
552 L 2.291
553 S 2.455
554 S 0.945
555 V -0.161
556 S 3.317
557 P 3.619
558 A 0.146
559 V 0.468
560 S -1.000 *
561 K -1.000 *
562 A -1.000 *
563 S -1.000 *
564 S -1.000 *
565 R -1.000 *
566 R -1.000 *
567 S -1.000 *
568 S -1.000 *
569 F -1.000 *
570 S -1.000 *
571 M -1.000 *
572 E -1.000 *
573 E -1.000 *
574 E -1.000 *
575 S -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 376.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MSHAAEPARD GVEASAEGPR AVFVLLEERR PADSAQLLSL NSLLPESGIV ADIELENVLD
70 80 90 100 110 120
PDSFYELKSQ PLPLRSSLPI SLQATPATPA TLSASSSAGG SRTPAMSSSS SSRVLLRQQL
130 140 150 160 170 180
MRAQAQEQER RERREQAAAA PFPSPAPASP AISVVGVSAG GHTLSRPPPA QVPREVLKVQ
190 200 210 220 230 240
THLENPTRYH LQQARRQQVK QYLSTTLGPK LASQALTPPP GPASAQPLPA PEAAHTTGPT
250 260 270 280 290 300
GSAPNSPMAL LTIGSSSEKE IDDVIDEIIS LESSYNDEML SYLPGGTTGL QLPSTLPVSG
310 320 330 340 350 360
NLLDVYSSQG VATPAITVSN SCPAELPNIK REISETEAKA LLKERQKKDN HNLIERRRRF
370 380 390 400 410 420
NINDRIKELG TLIPKSSDPE MRWNKGTILK ASVDYIRKLQ KEQQRSKDLE SRQRSLEQAN
430 440 450 460 470 480
RSLQLRIQEL ELQAQIHGLP VPPTPGLLSL ATTSASDSLK PEQLDIEEEG RPGAATFHVG
490 500 510 520 530 540
GGPAQNAPHQ QPPAPPSDAL LDLHFPSDHL GDLGDPFHLG LEDILMEEEE GVVGGLSGGA
550 560 570
LSPLRAASDP LLSSVSPAVS KASSRRSSFS MEEES
TF3E is a basic helix-loop-helix domain-containing transcription factor of the MiT/TFE family. This NES was identified by in silico prediction methods in Sendino et al. using possible XPO1/CRM1-cancer exportome proteins identified previously as putative CRM1-binders by Kirli et al. (not all proteins were validated in full-length). This putative NES peptide was tested using Rev(1.4)-GFP assay which showed weak activity. However, when tested in SRV
B/A reporter and co-expressed with CRM1, no cytoplasmic relocalization was observed, therefore CRM1-dependency was not demonstrated. This NES is predicted to be part of a leucine-zipper.
Ref.1Using a Simple Cellular Assay to Map NES Motifs in Cancer-Related Proteins, Gain Insight into CRM1-Mediated NES Export, and Search for NES-Harboring Micropeptides, Sendino et al., Int J Mol Sci, 2020 Ref.2A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning, Kirli et al., eLife, 2015
[1]. "Using a Simple Cellular Assay to Map NES Motifs in Cancer-Related Proteins, Gain Insight into CRM1-Mediated NES Export, and Search for NES-Harboring Micropeptides"
Sendino M, Omaetxebarria MJ, Prieto G, Rodriguez JA. (2020)
Int J Mol Sci,
21(17):E6341
PubMed[2]. "A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning"
Kırlı K, Karaca S, Dehne HJ, Samwer M, Pan KT, Lenz C, Urlaub H, Görlich D. (2015)
eLife,
4:e11466
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
