Show FASTA Format
>sp|Q14141|SEPT6_HUMAN Septin-6 OS=Homo sapiens OX=9606 GN=SEPTIN6 PE=1 SV=4
MAATDIARQVGEGCRTVPLAGHVGFDSLPDQLVNKSVSQGFCFNILCVGETGLGKSTLMD
TLFNTKFEGEPATHTQPGVQLQSNTYDLQESNVRLKLTIVSTVGFGDQINKEDSYKPIVE
FIDAQFEAYLQEELKIRRVLHTYHDSRIHVCLYFIAPTGHSLKSLDLVTMKKLDSKVNII
PIIAKADAISKSELTKFKIKITSELVSNGVQIYQFPTDDESVAEINGTMNAHLPFAVIGS
TEELKIGNKMMRARQYPWGTVQVENEAHCDFVKLREMLIRVNMEDLREQTHTRHYELYRR
CKLEEMGFKDTDPDSKPFSLQETYEAKRNEFLGELQKKEEEMRQMFVQRVKEKEAELKEA
EKELHEKFDRLKKLHQDEKKKLEDKKKSLDDEVNAFKQRKTAAELLQSQGSQAGGSQTLK
RDKEKKNNPWLCTE
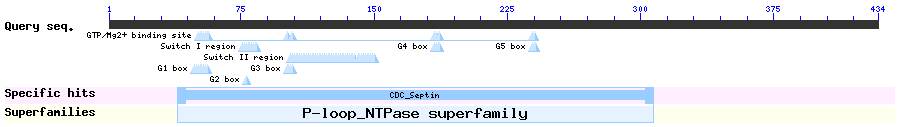
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 921168876079810230374338999725787420244112458863146885355644
Pred: CCCHHHHHHCCCCCEEEECCCCCCCCCCHHHHHHHCCCCCEEEEEEEECCCCCCHHHHHH
AA: MAATDIARQVGEGCRTVPLAGHVGFDSLPDQLVNKSVSQGFCFNILCVGETGLGKSTLMD
10 20 30 40 50 60
Conf: 210368999988888886011033100223303589999730125755687687613589
Pred: HHCCCCCCCCCCCCCCCCCEECCCCCCCCCCEEEEEEEEEEECCCCCCCCCCCCCCHHHH
AA: TLFNTKFEGEPATHTQPGVQLQSNTYDLQESNVRLKLTIVSTVGFGDQINKEDSYKPIVE
70 80 90 100 110 120
Conf: 489999998898761413103235661179999866899875422002210245643223
Pred: HHHHHHHHHHHHHHHHHHHHCCCCCCCEEEEEEEECCCCCCCCCCCHHHHCCCCCCCCEE
AA: FIDAQFEAYLQEELKIRRVLHTYHDSRIHVCLYFIAPTGHSLKSLDLVTMKKLDSKVNII
130 140 150 160 170 180
Conf: 333126645777887799999888652690888657990458987410146654358504
Pred: EEEECCCCCCHHHHHHHHHHHHHHHHHCCEEEEECCCCHHHHHHHHHHCCCCCCEEEEEC
AA: PIIAKADAISKSELTKFKIKITSELVSNGVQIYQFPTDDESVAEINGTMNAHLPFAVIGS
190 200 210 220 230 240
Conf: 531121320001113670358842588761365576775516035565432346898853
Pred: CCCEEECCCCHHCCCCCCEEEEECCCCCCCHHHHHHHHHHCCHHHHHHHHHHHHHHHHHH
AA: TEELKIGNKMMRARQYPWGTVQVENEAHCDFVKLREMLIRVNMEDLREQTHTRHYELYRR
250 260 270 280 290 300
Conf: 011105977789999953348989998544455633249999999888777788888898
Pred: HCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH
AA: CKLEEMGFKDTDPDSKPFSLQETYEAKRNEFLGELQKKEEEMRQMFVQRVKEKEAELKEA
310 320 330 340 350 360
Conf: 898876555543234454444433100025677775432248999742256778864311
Pred: HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCC
AA: EKELHEKFDRLKKLHQDEKKKLEDKKKSLDDEVNAFKQRKTAAELLQSQGSQAGGSQTLK
370 380 390 400 410 420
Conf: 00012489986469
Pred: CCCCCCCCCCCCCC
AA: RDKEKKNNPWLCTE
430
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 A -1.000 *
4 T -1.000 *
5 D -1.000 *
6 I -1.000 *
7 A -1.000 *
8 R -1.000 *
9 Q -1.000 *
10 V -1.000 *
11 G -1.000 *
12 E -1.156
13 G -1.002
14 C -1.162
15 R -0.860
16 T -1.021
17 V -0.819
18 P -0.857
19 L -0.235
20 A -1.146
21 G 0.262
22 H -0.477
23 V 0.888
24 G 3.077
25 F 0.986
26 D -0.127
27 S 0.007
28 L 1.138
29 P 1.794
30 D -0.424
31 Q 2.286
32 L -0.468
33 V -0.078
34 N -0.223
35 K 0.359
36 S -0.214
37 V -0.104
38 S -0.363
39 Q -0.509
40 G 1.657
41 F 1.027
42 C -1.061
43 F 1.871
44 N 0.975
45 I 0.469
46 L 1.783
47 C 0.829
48 V 0.698
49 G 2.590
50 E 0.397
51 T 0.393
52 G 1.550
53 L 0.346
54 G 1.984
55 K 1.984
56 S 0.885
57 T 1.594
58 L 1.400
59 M 0.750
60 D 0.239
61 T 0.996
62 L 1.234
63 F 2.396
64 N -0.712
65 T -0.455
66 K -0.810
67 F -0.680
68 E -0.876
69 G -0.974
70 E -1.129
71 P -0.801
72 A -0.996
73 T -1.078
74 H -0.788
75 T -1.260
76 Q -1.156
77 P -0.542
78 G -0.822
79 V 0.511
80 Q -0.926
81 L 0.854
82 Q -0.706
83 S -0.687
84 N -0.880
85 T -0.525
86 Y -0.509
87 D -0.802
88 L 1.037
89 Q -0.649
90 E 2.604
91 S -0.354
92 N -0.616
93 V 1.333
94 R -0.735
95 L 0.832
96 K -0.603
97 L 1.612
98 T 0.211
99 I 0.538
100 V 0.811
101 S 0.003
102 T 1.280
103 V -0.339
104 G 2.198
105 F 1.692
106 G 2.109
107 D 1.333
108 Q -0.513
109 I 0.935
110 N 1.219
111 K 0.344
112 E -0.715
113 D -0.613
114 S 0.342
115 Y -0.257
116 K -0.869
117 P -0.126
118 I 1.237
119 V -0.304
120 E -0.383
121 F -0.149
122 I 0.895
123 D 0.510
124 A -0.636
125 Q 0.705
126 F 1.004
127 E 0.102
128 A -0.900
129 Y 1.058
130 L 1.100
131 Q -0.745
132 E -0.350
133 E 2.621
134 L -0.595
135 K -0.267
136 I 0.005
137 R -0.911
138 R 1.079
139 V -0.942
140 L -1.092
141 H -1.144
142 T -1.109
143 Y -0.686
144 H -0.911
145 D 2.616
146 S -0.664
147 R 2.088
148 I 1.010
149 H 1.851
150 V 0.365
151 C 1.134
152 L 0.924
153 Y 2.195
154 F 1.286
155 I 1.274
156 A -0.559
157 P 1.489
158 T -0.366
159 G 1.326
160 H 1.055
161 S -0.369
162 L 0.917
163 K 0.278
164 S -0.187
165 L 0.816
166 D 1.622
167 L 0.823
168 V -0.668
169 T -0.013
170 M 1.285
171 K 0.194
172 K -0.841
173 L 1.298
174 D -0.088
175 S -0.667
176 K 0.472
177 V 0.950
178 N 1.458
179 I 0.891
180 I 1.753
181 P 1.229
182 I 0.929
183 I 0.993
184 A 1.013
185 K 1.398
186 A 1.583
187 D 2.167
188 A 0.102
189 I 0.414
190 S 0.276
191 K -0.921
192 S -1.004
193 E 0.612
194 L 0.164
195 T -1.214
196 K -1.080
197 F 0.327
198 K 2.030
199 I -1.206
200 K -0.281
201 I 1.045
202 T -0.329
203 S -0.890
204 E 0.096
205 L 0.634
206 V -1.019
207 S -1.114
208 N -0.127
209 G -0.334
210 V 1.121
211 Q -0.352
212 I 0.323
213 Y 1.067
214 Q -0.839
215 F -0.510
216 P -0.257
217 T -0.889
218 D -0.852
219 D -0.407
220 E -0.903
221 S -0.612
222 V -1.003
223 A -0.914
224 E -0.791
225 I -1.015
226 N -0.805
227 G -1.000
228 T -1.002
229 M -0.256
230 N -1.040
231 A -0.727
232 H -1.046
233 L -0.083
234 P 2.017
235 F 0.391
236 A 1.102
237 V 0.965
238 I 0.874
239 G 1.497
240 S 0.530
241 T -0.836
242 E -1.001
243 E -0.793
244 L -0.261
245 K -0.752
246 I -0.976
247 G -0.432
248 N 0.122
249 K -0.357
250 M -0.906
251 M -0.206
252 R 0.608
253 A 0.574
254 R 2.634
255 Q -0.866
256 Y 1.413
257 P 0.109
258 W 2.246
259 G 2.275
260 T 0.166
261 V 0.704
262 Q 0.231
263 V 0.337
264 E 0.873
265 N 0.911
266 E -0.683
267 A -0.714
268 H 2.067
269 C 0.706
270 D 1.093
271 F 0.622
272 V -0.561
273 K -0.700
274 L 1.875
275 R 0.614
276 E -0.691
277 M -0.034
278 L 0.785
279 I 0.570
280 R -0.069
281 V -0.315
282 N 0.320
283 M 0.876
284 E -0.050
285 D 0.551
286 L 1.878
287 R 0.329
288 E -0.044
289 Q -0.696
290 T 2.227
291 H -0.389
292 T -1.105
293 R -1.010
294 H 0.813
295 Y 1.669
296 E 1.444
297 L -0.936
298 Y 0.977
299 R 2.226
300 R -0.772
301 C -1.125
302 K -0.177
303 L 0.819
304 E -0.818
305 E -0.755
306 M -0.863
307 G -0.873
308 F -1.220
309 K -1.154
310 D -0.879
311 T -1.075
312 D -0.978
313 P -0.969
314 D -1.017
315 S -1.191
316 K -1.004
317 P -0.892
318 F -1.135
319 S -0.927
320 L -1.199
321 Q -1.102
322 E -1.152
323 T -1.108
324 Y -1.237
325 E -0.918
326 A -1.055
327 K -1.042
328 R -0.972
329 N -1.118
330 E -1.035
331 F -1.068
332 L -1.106
333 G -1.124
334 E -0.841
335 L -0.172
336 Q -0.745
337 K -0.485
338 K -0.396
339 E 0.184
340 E -0.739
341 E -0.427
342 M -0.341
343 R -0.310
344 Q -0.981
345 M -0.892
346 F 0.180
347 V -0.823
348 Q -0.947
349 R 0.221
350 V -0.291
351 K -0.570
352 E -0.597
353 K -0.137
354 E -0.843
355 A -0.844
356 E -0.591
357 L 0.079
358 K -0.271
359 E -0.410
360 A -0.730
361 E 0.110
362 K -0.921
363 E -0.281
364 L -0.094
365 H -0.988
366 E -0.981
367 K -0.054
368 F -0.818
369 D 0.114
370 R -0.934
371 L -0.460
372 K -0.609
373 K -0.712
374 L -0.981
375 H -0.525
376 Q -0.659
377 D -0.733
378 E -0.427
379 K -1.103
380 K -0.971
381 K -0.897
382 L 0.098
383 E -0.323
384 D -0.624
385 K -0.513
386 K -0.554
387 K -0.835
388 S -0.988
389 L 0.121
390 D -0.069
391 D -0.822
392 E -0.155
393 V -0.592
394 N -1.103
395 A -0.987
396 F -0.735
397 K -1.137
398 Q -0.840
399 R -0.944
400 K -0.626
401 T -1.000 *
402 A -1.000 *
403 A -1.000 *
404 E -1.000 *
405 L -1.000 *
406 L -1.000 *
407 Q -1.000 *
408 S -1.000 *
409 Q -1.000 *
410 G -1.000 *
411 S -1.000 *
412 Q -1.000 *
413 A -1.000 *
414 G -1.000 *
415 G -1.000 *
416 S -1.000 *
417 Q -1.000 *
418 T -1.000 *
419 L -1.000 *
420 K -1.000 *
421 R -1.000 *
422 D -1.000 *
423 K -1.000 *
424 E -1.000 *
425 K -1.000 *
426 K -1.000 *
427 N -1.000 *
428 N -1.000 *
429 P -1.000 *
430 W -1.000 *
431 L -1.000 *
432 C -1.000 *
433 T -1.000 *
434 E -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 378.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MAATDIARQV GEGCRTVPLA GHVGFDSLPD QLVNKSVSQG FCFNILCVGE TGLGKSTLMD
70 80 90 100 110 120
TLFNTKFEGE PATHTQPGVQ LQSNTYDLQE SNVRLKLTIV STVGFGDQIN KEDSYKPIVE
130 140 150 160 170 180
FIDAQFEAYL QEELKIRRVL HTYHDSRIHV CLYFIAPTGH SLKSLDLVTM KKLDSKVNII
190 200 210 220 230 240
PIIAKADAIS KSELTKFKIK ITSELVSNGV QIYQFPTDDE SVAEINGTMN AHLPFAVIGS
250 260 270 280 290 300
TEELKIGNKM MRARQYPWGT VQVENEAHCD FVKLREMLIR VNMEDLREQT HTRHYELYRR
310 320 330 340 350 360
CKLEEMGFKD TDPDSKPFSL QETYEAKRNE FLGELQKKEE EMRQMFVQRV KEKEAELKEA
370 380 390 400 410 420
EKELHEKFDR LKKLHQDEKK KLEDKKKSLD DEVNAFKQRK TAAELLQSQG SQAGGSQTLK
430
RDKEKKNNPW LCTE
Septin-6 is filament-forming cytoskeletal GTPase required for the actin cytoskeleton. This NES was identified by in silico prediction methods in Sendino et al. using possible XPO1/CRM1-cancer exportome proteins identified previously as putative CRM1-binders by Kirli et al. (not all proteins were validated in full-length). This putative NES peptide was tested using Rev(1.4)-GFP assay which showed weak activity. It is not tested in SRV
B/A reporter with co-expression with CRM1, therefore no CRM1-dependency has been demonstrated. Putative NES is part of the domain and is in the interface of the trimeric X-Ray structure.
Ref.1Using a Simple Cellular Assay to Map NES Motifs in Cancer-Related Proteins, Gain Insight into CRM1-Mediated NES Export, and Search for NES-Harboring Micropeptides, Sendino et al., Int J Mol Sci, 2020 Ref.2A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning, Kirli et al., eLife, 2015
[1]. "Using a Simple Cellular Assay to Map NES Motifs in Cancer-Related Proteins, Gain Insight into CRM1-Mediated NES Export, and Search for NES-Harboring Micropeptides"
Sendino M, Omaetxebarria MJ, Prieto G, Rodriguez JA. (2020)
Int J Mol Sci,
21(17):E6341
PubMed[2]. "A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning"
Kırlı K, Karaca S, Dehne HJ, Samwer M, Pan KT, Lenz C, Urlaub H, Görlich D. (2015)
eLife,
4:e11466
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
