Show FASTA Format
>gi|239938878|sp|Q62074.3|KPCI_MOUSE RecName: Full=Protein kinase C iota type; AltName: Full=Atypical protein kinase C-lambda/iota; Short=aPKC-lambda/iota; AltName: Full=nPKC-iota
MPTQRDSSTMSHTVACGGGGDHSHQVRVKAYYRGDIMITHFEPSISFEGLCSEVRDMCSFDNEQPFTMKW
IDEEGDPCTVSSQLELEEAFRLYELNKDSELLIHVFPCVPERPGMPCPGEDKSIYRRGARRWRKLYCANG
HTFQAKRFNRRAHCAICTDRIWGLGRQGYKCINCKLLVHKKCHKLVTIECGRHSLPPEPMMPMDQTMHPD
HTQTVIPYNPSSHESLDQVGEEKEAMNTRESGKASSSLGLQDFDLLRVIGRGSYAKVLLVRLKKTDRIYA
MKVVKKELVNDDEDIDWVQTEKHVFEQASNHPFLVGLHSCFQTESRLFFVIEYVNGGDLMFHMQRQRKLP
EEHARFYSAEISLALNYLHERGIIYRDLKLDNVLLDSEGHIKLTDYGMCKEGLRPGDTTSTFCGTPNYIA
PEILRGEDYGFSVDWWALGVLMFEMMAGRSPFDIVGSSDNPDQNTEDYLFQVILEKQIRIPRSLSVKAAS
VLKSFLNKDPKERLGCHPQTGFADIQGHPFFRNVDWDMMEQKQVVPPFKPNISGEFGLDNFDSQFTNEPV
QLTPDDDDIVRKIDQSEFEGFEYINPLLMSAEECV
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 997789986322111699998774168988633964873048988957777998643279
Pred: CCCCCCCCCCCCCCCCCCCCCCCCEEEEEEEECCCEEEEECCCCCCHHHHHHHHHHHCCC
AA: MPTQRDSSTMSHTVACGGGGDHSHQVRVKAYYRGDIMITHFEPSISFEGLCSEVRDMCSF
10 20 30 40 50 60
Conf: 999941001105789965457566699997532105775168741589999999999999
Pred: CCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCCCCCEEEEECCCCCCCCCCCCCCC
AA: DNEQPFTMKWIDEEGDPCTVSSQLELEEAFRLYELNKDSELLIHVFPCVPERPGMPCPGE
70 80 90 100 110 120
Conf: 863110234342000541593334323075322333332001122452222200002440
Pred: CCCCCCCCCCCCCCCEEECCCEEEEEECCCCCCCCCCCCCHHHCCCCCCCCCCEEEEEEE
AA: DKSIYRRGARRWRKLYCANGHTFQAKRFNRRAHCAICTDRIWGLGRQGYKCINCKLLVHK
130 140 150 160 170 180
Conf: 344453344678789999899999999999976545689999887555664212467556
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: KCHKLVTIECGRHSLPPEPMMPMDQTMHPDHTQTVIPYNPSSHESLDQVGEEKEAMNTRE
190 200 210 220 230 240
Conf: 788888879898740899504556448999872588257887544100368886420122
Pred: CCCCCCCCCCCCCCEEEEEECCCCCEEEEEEEECCCCEEEEEEEEECCCCCCCCCCHHHH
AA: SGKASSSLGLQDFDLLRVIGRGSYAKVLLVRLKKTDRIYAMKVVKKELVNDDEDIDWVQT
250 260 270 280 290 300
Conf: 568997732998231012200158704899997407822566777304984442142256
Pred: HHHHHHHHCCCCCEECCCCCCCCCCEEEEEEEEEECCCHHHHHHHHHCCCCCCCCEEHHH
AA: EKHVFEQASNHPFLVGLHSCFQTESRLFFVIEYVNGGDLMFHMQRQRKLPEEHARFYSAE
310 320 330 340 350 360
Conf: 899986431025110017655022168998010024431279999986443468987520
Pred: HHHHHHHHHHCCCEEECCCCCCEEECCCCCEEECCCCCCCCCCCCCCCCCCCCCCCCCCH
AA: ISLALNYLHERGIIYRDLKLDNVLLDSEGHIKLTDYGMCKEGLRPGDTTSTFCGTPNYIA
370 380 390 400 410 420
Conf: 112037987653114568999999850999975479999999995568899974126212
Pred: HHHHCCCCCCCCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCHHHHHHHHHHCCCCC
AA: PEILRGEDYGFSVDWWALGVLMFEMMAGRSPFDIVGSSDNPDQNTEDYLFQVILEKQIRI
430 440 450 460 470 480
Conf: 643206478998775239988736878998721001498954556343312447999988
Pred: CCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHCCCCCCCCCC
AA: PRSLSVKAASVLKSFLNKDPKERLGCHPQTGFADIQGHPFFRNVDWDMMEQKQVVPPFKP
490 500 510 520 530 540
Conf: 7788889889998887899612789746762038575588411197676833459
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHCCCCCCCCCCEECCCCCCCCCCCC
AA: NISGEFGLDNFDSQFTNEPVQLTPDDDDIVRKIDQSEFEGFEYINPLLMSAEECV
550 560 570 580 590
Show Conservation Score by AL2CO
1 M -1.000 *
2 P -1.000 *
3 T -1.000 *
4 Q -1.000 *
5 R -1.000 *
6 D -1.000 *
7 S -1.000 *
8 S -1.000 *
9 T -1.000 *
10 M -1.000 *
11 S -1.000 *
12 H -1.000 *
13 T -1.000 *
14 V -1.000 *
15 A -1.000 *
16 C -1.000 *
17 G -1.000 *
18 G -1.000 *
19 G -1.000 *
20 G -1.000 *
21 D -1.000 *
22 H -1.000 *
23 S -1.000 *
24 H -1.000 *
25 Q -1.000 *
26 V -1.000 *
27 R -1.000 *
28 V -1.000 *
29 K -1.000 *
30 A -1.000 *
31 Y -1.000 *
32 Y -1.000 *
33 R -1.000 *
34 G -1.000 *
35 D -1.000 *
36 I -1.000 *
37 M -1.000 *
38 I -1.000 *
39 T -1.000 *
40 H -1.000 *
41 F -1.000 *
42 E -1.000 *
43 P -1.000 *
44 S -1.000 *
45 I -1.000 *
46 S -1.000 *
47 F -1.000 *
48 E -1.000 *
49 G -1.000 *
50 L -1.000 *
51 C -1.000 *
52 S -1.000 *
53 E -1.000 *
54 V -1.000 *
55 R -1.000 *
56 D -1.000 *
57 M -1.000 *
58 C -1.000 *
59 S -1.000 *
60 F -1.000 *
61 D -1.000 *
62 N -1.000 *
63 E -1.000 *
64 Q -1.000 *
65 P -1.000 *
66 F -1.000 *
67 T -1.000 *
68 M -1.000 *
69 K -1.000 *
70 W -1.000 *
71 I -1.000 *
72 D -1.000 *
73 E -1.000 *
74 E -1.000 *
75 G -1.000 *
76 D -1.000 *
77 P -1.000 *
78 C -1.000 *
79 T -1.000 *
80 V -1.000 *
81 S -1.000 *
82 S -1.000 *
83 Q -1.000 *
84 L -1.000 *
85 E -1.000 *
86 L -1.000 *
87 E -1.000 *
88 E -1.000 *
89 A -1.000 *
90 F -1.000 *
91 R -1.000 *
92 L -1.000 *
93 Y -1.000 *
94 E -1.000 *
95 L -1.000 *
96 N -1.000 *
97 K -1.000 *
98 D -1.000 *
99 S -1.000 *
100 E -1.000 *
101 L -1.000 *
102 L -1.000 *
103 I -1.000 *
104 H -1.000 *
105 V -1.000 *
106 F -1.000 *
107 P -1.000 *
108 C -1.000 *
109 V -1.000 *
110 P -1.000 *
111 E -1.000 *
112 R -1.000 *
113 P -1.000 *
114 G -1.000 *
115 M -1.000 *
116 P -1.000 *
117 C -1.000 *
118 P -1.000 *
119 G -1.000 *
120 E -1.000 *
121 D -1.000 *
122 K -1.000 *
123 S -1.000 *
124 I -1.000 *
125 Y -1.000 *
126 R -1.000 *
127 R -1.000 *
128 G -1.000 *
129 A -1.000 *
130 R -1.000 *
131 R -1.000 *
132 W -1.000 *
133 R -1.000 *
134 K -1.000 *
135 L -0.029
136 Y -0.245
137 C -0.784
138 A -1.000 *
139 N -1.000 *
140 G -0.201
141 H 1.046
142 T -0.634
143 F 1.756
144 Q -1.003
145 A -0.861
146 K -0.004
147 R -0.468
148 F 0.396
149 N -0.473
150 R -0.374
151 R -0.133
152 A 0.096
153 H -0.710
154 C 1.795
155 A -0.655
156 I -0.163
157 C 1.883
158 T -0.618
159 D -0.112
160 R -0.442
161 I 0.400
162 W -0.189
163 G 0.292
164 L 0.124
165 G -0.602
166 R -0.545
167 Q 0.116
168 G -0.077
169 Y -0.619
170 K -0.455
171 C 0.139
172 I -0.703
173 N -0.863
174 C 0.198
175 K -0.680
176 L -0.509
177 L -0.419
178 V -0.541
179 H -0.175
180 K -0.727
181 K -0.644
182 C -0.584
183 H -0.773
184 K -0.926
185 L -0.979
186 V -0.831
187 T -0.969
188 I -1.146
189 E -1.110
190 C -0.878
191 G -1.085
192 R -0.976
193 H -1.228
194 S -1.127
195 L -0.954
196 P -1.142
197 P -1.074
198 E -0.975
199 P -1.159
200 M -1.029
201 M -1.118
202 P -0.982
203 M -1.018
204 D -1.111
205 Q -1.065
206 T -1.026
207 M -1.135
208 H -1.115
209 P -1.107
210 D -1.139
211 H -1.221
212 T -1.266
213 Q -1.166
214 T -1.248
215 V -1.215
216 I -1.151
217 P -1.199
218 Y -1.287
219 N -1.200
220 P -1.152
221 S -1.160
222 S -1.163
223 H -1.214
224 E -1.161
225 S -1.158
226 L -1.159
227 D -1.168
228 Q -1.247
229 V -1.188
230 G -1.245
231 E -1.186
232 E -1.192
233 K -1.231
234 E -1.256
235 A -1.220
236 M -1.217
237 N -1.162
238 T -1.269
239 R -1.178
240 E -1.186
241 S -1.163
242 G -1.188
243 K -1.186
244 A -1.061
245 S -1.015
246 S -1.146
247 S -0.932
248 L -0.781
249 G -0.799
250 L -0.586
251 Q -0.797
252 D -0.380
253 F 0.633
254 D -0.823
255 L 0.190
256 L -0.168
257 R -0.541
258 V 0.154
259 I 1.140
260 G 2.274
261 R 0.567
262 G 1.905
263 S 0.409
264 Y 1.102
265 A 1.011
266 K 1.470
267 V 1.750
268 L -0.104
269 L 0.902
270 V 0.529
271 R 0.162
272 L -0.899
273 K -0.100
274 K -0.944
275 T -0.734
276 D -0.543
277 R -0.978
278 I -0.431
279 Y 0.324
280 A 1.220
281 M 0.579
282 K 2.258
283 V -0.055
284 V 0.547
285 K 0.095
286 K 2.276
287 E -0.589
288 L -0.780
289 V 0.645
290 N -0.652
291 D -0.620
292 D -0.054
293 E -0.039
294 D 0.524
295 I -0.095
296 D -0.446
297 W 0.324
298 V 0.672
299 Q -0.379
300 T -0.363
301 E 1.894
302 K 0.835
303 H -0.370
304 V 1.533
305 F 1.198
306 E -0.594
307 Q -0.696
308 A -0.881
309 S -0.905
310 N -0.660
311 H 0.062
312 P 0.967
313 F 1.336
314 L 1.014
315 V 0.374
316 G -0.773
317 L 0.944
318 H -0.555
319 S 0.044
320 C 0.443
321 F 1.236
322 Q 0.889
323 T 1.094
324 E -0.948
325 S -0.232
326 R 0.441
327 L 0.719
328 F 0.262
329 F 0.854
330 V 1.317
331 I 0.771
332 E 0.619
333 Y 1.412
334 V -0.046
335 N -0.130
336 G 2.710
337 G 2.710
338 D 1.184
339 L 1.760
340 M 1.087
341 F -0.194
342 H 0.822
343 M 0.837
344 Q -0.185
345 R -0.680
346 Q -0.879
347 R -0.430
348 K -0.701
349 L 1.118
350 P -0.536
351 E -0.602
352 E -0.998
353 H -0.010
354 A 0.611
355 R 0.016
356 F 1.349
357 Y 2.264
358 S -0.025
359 A 0.547
360 E 1.578
361 I 0.841
362 S -0.178
363 L 0.004
364 A 1.354
365 L 0.791
366 N -0.926
367 Y 0.745
368 L 1.135
369 H 2.245
370 E -0.735
371 R -0.804
372 G -0.421
373 I 1.026
374 I 0.850
375 Y 1.245
376 R 2.264
377 D 1.084
378 L 0.895
379 K 2.264
380 L 0.993
381 D 1.507
382 N 2.264
383 V 1.075
384 L 1.285
385 L 0.862
386 D -0.004
387 S -1.085
388 E -0.689
389 G 1.608
390 H 1.028
391 I 0.832
392 K 0.075
393 L 0.845
394 T 0.714
395 D 2.182
396 Y 1.300
397 G 1.032
398 M 0.669
399 C 1.294
400 K 1.520
401 E -0.136
402 G -0.040
403 L 0.599
404 R -1.228
405 P -1.025
406 G -0.615
407 D -0.954
408 T -0.687
409 T 0.628
410 S -0.594
411 T 1.957
412 F 1.454
413 C 1.272
414 G 1.881
415 T 1.425
416 P 0.388
417 N 0.680
418 Y 0.825
419 I 0.814
420 A 1.633
421 P 1.919
422 E 1.461
423 I 1.021
424 L 0.530
425 R -0.753
426 G -0.375
427 E -0.782
428 D -0.748
429 Y 1.529
430 G 0.116
431 F -0.896
432 S -0.353
433 V 1.285
434 D 1.917
435 W 1.360
436 W 1.876
437 A 0.402
438 L 0.370
439 G 1.831
440 V 0.505
441 L 0.782
442 M 0.063
443 F 1.515
444 E 1.004
445 M 1.465
446 M 0.580
447 A -0.397
448 G 2.040
449 R -0.238
450 S 0.473
451 P 1.768
452 F 1.271
453 D -0.616
454 I -0.554
455 V -1.000 *
456 G -1.000 *
457 S -1.000 *
458 S -1.000 *
459 D -1.000 *
460 N -1.000 *
461 P -1.000 *
462 D -1.000 *
463 Q -0.226
464 N -0.405
465 T 0.299
466 E -0.430
467 D -0.053
468 Y -0.800
469 L 0.654
470 F 0.895
471 Q -0.669
472 V -0.644
473 I 1.257
474 L -0.181
475 E -1.029
476 K -0.537
477 Q -0.861
478 I 0.293
479 R -1.047
480 I 0.386
481 P 0.222
482 R -0.873
483 S -1.004
484 L 0.215
485 S -0.496
486 V -0.909
487 K -0.714
488 A 0.574
489 A -0.855
490 S -0.685
491 V 0.215
492 L 0.752
493 K -0.758
494 S -0.312
495 F 1.079
496 L 1.173
497 N -0.822
498 K 1.281
499 D -0.443
500 P -0.261
501 K -0.915
502 E -0.849
503 R 2.710
504 L 1.648
505 G 1.681
506 C -0.138
507 H -0.994
508 P -0.701
509 Q -1.032
510 T -0.851
511 G -0.198
512 F -0.975
513 A -1.025
514 D -0.614
515 I 0.980
516 Q -0.443
517 G -1.102
518 H 0.842
519 P -0.990
520 F 1.418
521 F 1.690
522 R -0.947
523 N -1.061
524 V 0.566
525 D -0.446
526 W 1.709
527 D -0.992
528 M -0.662
529 M 0.666
530 E -1.021
531 Q -0.873
532 K -0.066
533 Q -0.436
534 V 0.043
535 V -0.814
536 P 1.018
537 P 0.635
538 F 0.800
539 K -0.534
540 P 2.317
541 N -0.907
542 I -0.437
543 S -0.867
544 G -0.282
545 E -0.922
546 F -1.094
547 G 0.191
548 L -0.623
549 D -0.868
550 N 0.412
551 F 1.438
552 D 1.034
553 S -0.823
554 Q -0.598
555 F 2.016
556 T 0.929
557 N -0.881
558 E -0.387
559 P -0.762
560 V 0.247
561 Q -1.071
562 L -0.681
563 T 0.793
564 P -0.680
565 D -0.666
566 D -0.686
567 D -0.854
568 D -1.098
569 I -1.033
570 V -0.523
571 R -0.934
572 K -1.016
573 I -0.899
574 D -0.846
575 Q -0.336
576 S -0.998
577 E -0.941
578 F 2.295
579 E -0.889
580 G 0.087
581 F 1.567
582 E 0.813
583 Y 0.435
584 I -0.306
585 N -0.004
586 P -0.113
587 L -1.072
588 L -1.000 *
589 M -1.000 *
590 S -1.000 *
591 A -1.000 *
592 E -1.000 *
593 E -1.000 *
594 C -1.000 *
595 V -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 42.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MPTQRDSSTM SHTVACGGGG DHSHQVRVKA YYRGDIMITH FEPSISFEGL CSEVRDMCSF
70 80 90 100 110 120
DNEQPFTMKW IDEEGDPCTV SSQLELEEAF RLYELNKDSE LLIHVFPCVP ERPGMPCPGE
130 140 150 160 170 180
DKSIYRRGAR RWRKLYCANG HTFQAKRFNR RAHCAICTDR IWGLGRQGYK CINCKLLVHK
190 200 210 220 230 240
KCHKLVTIEC GRHSLPPEPM MPMDQTMHPD HTQTVIPYNP SSHESLDQVG EEKEAMNTRE
250 260 270 280 290 300
SGKASSSLGL QDFDLLRVIG RGSYAKVLLV RLKKTDRIYA MKVVKKELVN DDEDIDWVQT
310 320 330 340 350 360
EKHVFEQASN HPFLVGLHSC FQTESRLFFV IEYVNGGDLM FHMQRQRKLP EEHARFYSAE
370 380 390 400 410 420
ISLALNYLHE RGIIYRDLKL DNVLLDSEGH IKLTDYGMCK EGLRPGDTTS TFCGTPNYIA
430 440 450 460 470 480
PEILRGEDYG FSVDWWALGV LMFEMMAGRS PFDIVGSSDN PDQNTEDYLF QVILEKQIRI
490 500 510 520 530 540
PRSLSVKAAS VLKSFLNKDP KERLGCHPQT GFADIQGHPF FRNVDWDMME QKQVVPPFKP
550 560 570 580 590
NISGEFGLDN FDSQFTNEPV QLTPDDDDIV RKIDQSEFEG FEYINPLLMS AEECV
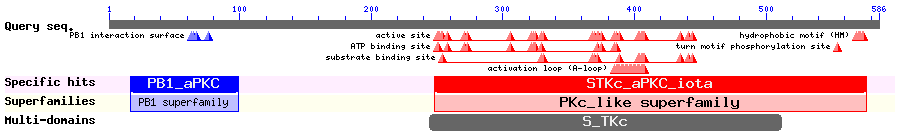
λPKC belongs to atypical protein kinase C family. λPKC has a single zinc finger domain (in comparison with two in other types of PKCs) regulated by lipid products such as ceramide. Perander et al. found that λPKC is mainly located in cytoplasm. But mutations in the ATP-binding site (like K273/W) will cause the protein to be nuclear, indicating the presence of NLS in the sequence. Fragments of λPKC, fused to GFP were examined for their localization. It was found that GFP-λPKC(132-185) is exclusively nuclear, suggesting that it contains a functional NLS. On the other hand, GFP-λPKC(185-251) is exported from the nucleus. LMB treatment induces nuclear accumulation of λPKC. F244A/L246A mutation cause the whole protein to become nuclear also. Note that there is a 9 aa difference between the numbering of the paper and SWISSPROT.
[1]. "Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase C λ"
Perander, M., Bjorkoy, G., Johansen, T. (2001)
J Biol Chem,
276:13015-13024
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
