Summary for Zinc finger protein RFP (NES ID: 43)
Full Name
Zinc finger protein RFP
UniProt Alternative Names
Ret finger protein
Tripartite motif-containing protein 27
RING finger protein 76
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
*shown as underlined residues in the full sequence
132LEEAVEGFKEQIQNQLDHLKRVKDLKKRRRAQGEQARAELLSLTQMEREKIVWEFEQLYH
SLKEHEYRLLARLEELDLAIYNSINGAITQFSCNISHLSSLIAQLEEKQQQPTRELLQDI
GDTL
255
Secondary Structure of Export Signal
Unknown
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|132517|sp|P14373.1|TRI27_HUMAN RecName: Full=Zinc finger protein RFP; AltName: Full=RING finger protein 76; AltName: Full=Ret finger protein; AltName: Full=Tripartite motif-containing protein 27
MASGSVAECLQQETTCPVCLQYFAEPMMLDCGHNICCACLARCWGTAETNVSCPQCRETFPQRHMRPNRH
LANVTQLVKQLRTERPSGPGGEMGVCEKHREPLKLYCEEDQMPICVVCDRSREHRGHSVLPLEEAVEGFK
EQIQNQLDHLKRVKDLKKRRRAQGEQARAELLSLTQMEREKIVWEFEQLYHSLKEHEYRLLARLEELDLA
IYNSINGAITQFSCNISHLSSLIAQLEEKQQQPTRELLQDIGDTLSRAERIRIPEPWITPPDLQEKIHIF
AQKCLFLTESLKQFTEKMQSDMEKIQELREAQLYSVDVTLDPDTAYPSLILSDNLRQVRYSYLQQDLPDN
PERFNLFPCVLGSPCFIAGRHYWEVEVGDKAKWTIGVCEDSVCRKGGVTSAPQNGFWAVSLWYGKEYWAL
TSPMTALPLRTPLQRVGIFLDYDAGEVSFYNVTERCHTFTFSHATFCGPVRPYFSLSYSGGKSAAPLIIC
PMSGIDGFSGHVGNHGHSMETSP
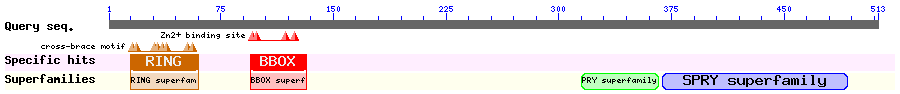
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 998630001222110422300005997456767876134651214799987862348468
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHCCCCCCCCCCCCCCCC
AA: MASGSVAECLQQETTCPVCLQYFAEPMMLDCGHNICCACLARCWGTAETNVSCPQCRETF
10 20 30 40 50 60
Conf: 899999984555699999851126999999898403431353335522588210223574
Pred: CCCCCCCCHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHCCCCCCCCCCCCCEEECCCCC
AA: PQRHMRPNRHLANVTQLVKQLRTERPSGPGGEMGVCEKHREPLKLYCEEDQMPICVVCDR
70 80 90 100 110 120
Conf: 457579842114556799899999988876999999999998568999999999999999
Pred: CCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH
AA: SREHRGHSVLPLEEAVEGFKEQIQNQLDHLKRVKDLKKRRRAQGEQARAELLSLTQMERE
130 140 150 160 170 180
Conf: 999999999976999999999998998699999975435557313568999999999991
Pred: HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH
AA: KIVWEFEQLYHSLKEHEYRLLARLEELDLAIYNSINGAITQFSCNISHLSSLIAQLEEKQ
190 200 210 220 230 240
Conf: 599688875356887521268779984349001112113421205999999973676666
Pred: CCCHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHH
AA: QQPTRELLQDIGDTLSRAERIRIPEPWITPPDLQEKIHIFAQKCLFLTESLKQFTEKMQS
250 260 270 280 290 300
Conf: 689999977412421027609788898631103888187266766899998997888921
Pred: HHHHHHHHHHHCCCCEEEECCCCCCCCCCCCCCCCCEEECCCCCCCCCCCCCCCCCCCEE
AA: DMEKIQELREAQLYSVDVTLDPDTAYPSLILSDNLRQVRYSYLQQDLPDNPERFNLFPCV
310 320 330 340 350 360
Conf: 027986676316999832988507997447776579966799982789998689938874
Pred: ECCCCCCCCCCEEEEEECCCCCEEEEEECCCCCCCCCCCCCCCCCEEEEEECCCCCEEEC
AA: LGSPCFIAGRHYWEVEVGDKAKWTIGVCEDSVCRKGGVTSAPQNGFWAVSLWYGKEYWAL
370 380 390 400 410 420
Conf: 679967899899985889954179803697569997033418999897531103527799
Pred: CCCCCCCCCCCCCCEEEEEEECCCCEEEEEECCCCCEEEEECCCCCCCCCCCCEECCCCC
AA: TSPMTALPLRTPLQRVGIFLDYDAGEVSFYNVTERCHTFTFSHATFCGPVRPYFSLSYSG
430 440 450 460 470 480
Conf: 987889153289998999898779887799999
Pred: CCCCCCEEECCCCCCCCCCCCCCCCCCCCCCCC
AA: GKSAAPLIICPMSGIDGFSGHVGNHGHSMETSP
490 500 510
Show Conservation Score by AL2CO
1 M 3.930
2 A 1.132
3 S -0.095
4 G -0.556
5 S -0.636
6 V -0.493
7 A 0.093
8 E -0.449
9 C -0.665
10 L 0.573
11 Q -0.575
12 Q -0.248
13 E 0.732
14 T 0.776
15 T -0.219
16 C 3.930
17 P 1.009
18 V 1.266
19 C 3.127
20 L 0.014
21 Q -0.054
22 Y -0.193
23 F 0.824
24 A -0.242
25 E 0.463
26 P 2.747
27 M 1.268
28 M 0.558
29 L 0.919
30 D -0.276
31 C 3.930
32 G 1.090
33 H 2.983
34 N 0.944
35 I 1.489
36 C 3.930
37 C 0.048
38 A -0.644
39 C 3.442
40 L 1.241
41 A -0.718
42 R -0.615
43 C -0.539
44 W -0.776
45 G -0.605
46 T -0.576
47 A -0.633
48 E -0.570
49 T -0.761
50 N -0.778
51 V -0.615
52 S -0.708
53 C 3.596
54 P 2.533
55 Q -0.230
56 C 3.930
57 R 0.384
58 E -0.570
59 T -0.758
60 F -0.057
61 P -0.634
62 Q -0.631
63 R -0.370
64 H -0.470
65 M 0.145
66 R -0.218
67 P -0.019
68 N 2.095
69 R -0.218
70 H -0.555
71 L 1.427
72 A 0.198
73 N 0.282
74 V 0.856
75 T 0.369
76 Q -0.196
77 L -0.391
78 V -0.028
79 K 0.732
80 Q -0.450
81 L -0.782
82 R -0.875
83 T -0.362
84 E -0.814
85 R -0.724
86 P -0.874
87 S -0.878
88 G -0.720
89 P -0.640
90 G -0.829
91 G -0.834
92 E -0.713
93 M -0.535
94 G -0.994
95 V -0.513
96 C 2.624
97 E -0.621
98 K -0.493
99 H 2.479
100 R -0.211
101 E 1.061
102 P -0.709
103 L 0.597
104 K -0.469
105 L 0.204
106 Y 1.013
107 C 1.836
108 E -0.277
109 E -0.264
110 D 0.698
111 Q -0.240
112 M -0.436
113 P -0.239
114 I 0.644
115 C 2.128
116 V -0.648
117 V -0.499
118 C 2.361
119 D -0.773
120 R -0.542
121 S -0.226
122 R -0.518
123 E -0.468
124 H 0.654
125 R -0.437
126 G -0.477
127 H 0.726
128 S -0.734
129 V -0.174
130 L -0.666
131 P -0.026
132 L 0.301
133 E -0.230
134 E -0.011
135 A 0.044
136 V -0.599
137 E -0.504
138 G -0.771
139 F -0.289
140 K 0.435
141 E -0.593
142 Q -0.352
143 I 0.222
144 Q -0.300
145 N -0.777
146 Q -0.848
147 L 0.073
148 D -0.748
149 H -0.919
150 L 0.379
151 K -0.234
152 R -0.385
153 V -0.622
154 K -0.535
155 D -0.523
156 L -0.607
157 K -0.548
158 K -0.695
159 R -0.598
160 R -0.481
161 R -0.544
162 A -0.777
163 Q -0.821
164 G -0.747
165 E -0.321
166 Q -0.652
167 A -0.332
168 R -0.712
169 A -0.633
170 E -0.741
171 L -0.665
172 L 0.080
173 S -0.485
174 L -0.562
175 T -0.049
176 Q -0.380
177 M -0.799
178 E -0.433
179 R 0.059
180 E -0.579
181 K -0.601
182 I -0.058
183 V -0.639
184 W -0.729
185 E 0.060
186 F 0.408
187 E -0.550
188 Q -0.396
189 L -0.070
190 Y -0.607
191 H -0.564
192 S -0.170
193 L 0.967
194 K -0.755
195 E -0.527
196 H 0.007
197 E 0.130
198 Y -0.408
199 R -0.869
200 L -0.456
201 L -0.065
202 A -0.581
203 R -0.570
204 L 0.613
205 E -0.077
206 E -0.340
207 L 0.018
208 D -0.523
209 L -0.157
210 A -0.468
211 I -0.493
212 Y -0.525
213 N -0.651
214 S -0.620
215 I -0.003
216 N -0.545
217 G -0.423
218 A -0.668
219 I -0.548
220 T -0.452
221 Q -0.758
222 F 0.197
223 S -0.624
224 C -0.431
225 N -0.140
226 I -0.548
227 S -0.691
228 H -0.755
229 L 0.138
230 S -0.571
231 S -0.681
232 L -0.379
233 I -0.039
234 A -0.751
235 Q -0.083
236 L 0.261
237 E -0.150
238 E -0.531
239 K -0.320
240 Q -0.734
241 Q -0.291
242 Q -0.540
243 P -0.313
244 T -0.475
245 R -0.827
246 E -0.600
247 L -0.259
248 L 0.096
249 Q -0.289
250 D -0.333
251 I -0.394
252 G -0.550
253 D -0.684
254 T -0.680
255 L -0.579
256 S -0.717
257 R -0.256
258 A -0.916
259 E -0.744
260 R -0.970
261 I -0.952
262 R -0.867
263 I -0.755
264 P -0.803
265 E -0.818
266 P -0.511
267 W -0.886
268 I -0.767
269 T -0.824
270 P -0.877
271 P -0.786
272 D -0.840
273 L -0.794
274 Q -0.865
275 E -0.775
276 K -0.955
277 I -0.879
278 H -0.941
279 I -0.939
280 F -0.823
281 A -0.826
282 Q -0.842
283 K -0.868
284 C -0.961
285 L -0.932
286 F -0.863
287 L -0.836
288 T -0.762
289 E -1.010
290 S -0.717
291 L -0.971
292 K -0.833
293 Q -0.855
294 F -0.830
295 T -0.865
296 E -0.823
297 K -0.990
298 M -0.636
299 Q -0.915
300 S -0.930
301 D -0.914
302 M -0.787
303 E -0.959
304 K -0.953
305 I -0.561
306 Q -0.922
307 E -0.561
308 L -0.684
309 R -0.755
310 E -0.686
311 A -0.659
312 Q -0.726
313 L -0.874
314 Y -0.714
315 S -0.533
316 V -0.351
317 D -0.579
318 V 0.790
319 T -0.145
320 L 1.613
321 D 2.806
322 P 0.361
323 D -0.316
324 T 1.819
325 A 1.672
326 Y 0.548
327 P 0.074
328 S -0.776
329 L 1.485
330 I -0.227
331 L 1.055
332 S 0.948
333 D -0.108
334 N 0.488
335 L 0.030
336 R 0.379
337 Q -0.156
338 V 0.689
339 R -0.430
340 Y -0.572
341 S -0.148
342 Y -0.879
343 L -0.763
344 Q -0.856
345 Q -0.426
346 D -0.787
347 L -0.274
348 P -0.112
349 D -0.783
350 N -0.097
351 P -0.017
352 E -0.097
353 R 1.017
354 F 1.517
355 N -0.616
356 L -0.767
357 F -0.734
358 P -0.430
359 C -0.314
360 V 1.604
361 L 2.720
362 G 2.489
363 S -0.356
364 P 0.032
365 C -0.341
366 F 1.879
367 I -0.444
368 A 0.441
369 G 3.096
370 R 1.206
371 H 0.090
372 Y 0.764
373 W 2.485
374 E 1.298
375 V 1.782
376 E -0.277
377 V 1.662
378 G -0.163
379 D -0.141
380 K 0.091
381 A -0.742
382 K -0.789
383 W 2.006
384 T 0.087
385 I 1.124
386 G 3.566
387 V 1.236
388 C 1.661
389 E -0.404
390 D 0.179
391 S 0.356
392 V 0.388
393 C -0.680
394 R 1.038
395 K 0.326
396 G -0.337
397 G -0.872
398 V -0.673
399 T -0.794
400 S -0.814
401 A -0.831
402 P 0.299
403 Q -0.424
404 N -0.662
405 G 1.217
406 F 0.259
407 W 1.639
408 A -0.079
409 V 1.031
410 S -0.542
411 L -0.085
412 W -0.761
413 Y -0.910
414 G -0.227
415 K -0.681
416 E -0.619
417 Y 0.446
418 W -0.855
419 A 0.220
420 L -0.650
421 T -0.175
422 S -0.646
423 P -0.496
424 M -0.850
425 T -0.548
426 A -0.618
427 L 0.789
428 P -0.744
429 L 0.319
430 R -0.863
431 T -0.655
432 P -0.699
433 L 1.505
434 Q -0.481
435 R -0.368
436 V 1.549
437 G 1.262
438 I 2.493
439 F 0.089
440 L 2.331
441 D 1.791
442 Y 1.204
443 D 0.603
444 A -0.405
445 G 0.606
446 E -0.563
447 V 2.180
448 S 1.046
449 F 2.792
450 Y 0.937
451 N 1.522
452 V 0.440
453 T -0.252
454 E -0.025
455 R -0.266
456 C -0.223
457 H 0.346
458 T 1.179
459 F 1.292
460 T 0.337
461 F 2.341
462 S 0.060
463 H -0.311
464 A -0.529
465 T -0.387
466 F 2.094
467 C -0.451
468 G 1.060
469 P -0.200
470 V 1.391
471 R 0.083
472 P 2.272
473 Y 0.953
474 F 2.152
475 S -0.257
476 L 0.806
477 S -0.199
478 Y -0.823
479 S -0.668
480 G -0.161
481 G -0.218
482 K -0.589
483 S 0.425
484 A -0.290
485 A -0.309
486 P 1.138
487 L 1.736
488 I -0.182
489 I 1.903
490 C 0.423
491 P 0.431
492 M 0.543
493 S -1.000 *
494 G -1.000 *
495 I -1.000 *
496 D -1.000 *
497 G -1.000 *
498 F -1.000 *
499 S -1.000 *
500 G -1.000 *
501 H -1.000 *
502 V -1.000 *
503 G -1.000 *
504 N -1.000 *
505 H -1.000 *
506 G -1.000 *
507 H -1.000 *
508 S -1.000 *
509 M -1.000 *
510 E -1.000 *
511 T -1.000 *
512 S -1.000 *
513 P -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 43.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MASGSVAECL QQETTCPVCL QYFAEPMMLD CGHNICCACL ARCWGTAETN VSCPQCRETF
70 80 90 100 110 120
PQRHMRPNRH LANVTQLVKQ LRTERPSGPG GEMGVCEKHR EPLKLYCEED QMPICVVCDR
130 140 150 160 170 180
SREHRGHSVL PLEEAVEGFK EQIQNQLDHL KRVKDLKKRR RAQGEQARAE LLSLTQMERE
190 200 210 220 230 240
KIVWEFEQLY HSLKEHEYRL LARLEELDLA IYNSINGAIT QFSCNISHLS SLIAQLEEKQ
250 260 270 280 290 300
QQPTRELLQD IGDTLSRAER IRIPEPWITP PDLQEKIHIF AQKCLFLTES LKQFTEKMQS
310 320 330 340 350 360
DMEKIQELRE AQLYSVDVTL DPDTAYPSLI LSDNLRQVRY SYLQQDLPDN PERFNLFPCV
370 380 390 400 410 420
LGSPCFIAGR HYWEVEVGDK AKWTIGVCED SVCRKGGVTS APQNGFWAVS LWYGKEYWAL
430 440 450 460 470 480
TSPMTALPLR TPLQRVGIFL DYDAGEVSFY NVTERCHTFT FSHATFCGPV RPYFSLSYSG
490 500 510
GKSAAPLIIC PMSGIDGFSG HVGNHGHSME TSP
3D Structures in PDB
Not Available
Comments
Ret finger protein (RFP) is a member of the RBCC (RING-B box-coiled-coil) family. It was found to be oncogenic after its RBCC domain was linked to other proteins by DNA rearrangement. The localization of RFP depends on the cell types. In NIH3T3 cell, it is predominantly is cytoplasm, whereas in HepG2 cells, it is localized in the nucleus. Harbers et al. used EGFP labeled deletion mutants to determine which domains of RFP affect its localization. And they found that the coiled coil region is required of cytoplasmic localization in NIH3T3 cells, and the localization can be changed by LMB treatment. In addition, they found that when fused, the coiled-coil region pf RFP can relocate the nuclear protein Gal4DBD to the cytoplasm. L204A/L207A/L209A mutation will block the relocation. As for the nuclear localization in HepG2 cell, they found that the NES is still functional. However, when full-length protein is fused, Gal4DBD is not exported, indicating that somehow the NES is masked. Protein kinase C is a possible candidate as a control for RFP shuttling.
References
[1]. "Intracellular localization of the Ret finger protein depends on a functional nuclear export signal and protein kinase C activation."
Harbers, M., Nomura, T., Ohno, S., Ishii, S. (2001)
J Biol Chem,
276:48596-48607
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
