Summary for Cdc25 (NES ID: 46)
Full Name
M-phase inducer phosphatase 1-B
UniProt Alternative Names
None
Organism
Xenopus laevis (African clawed frog)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
Undetermined
Secondary Structure of Export Signal
Unknown
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|266557|sp|P30309.1|MPI1B_XENLA RecName: Full=M-phase inducer phosphatase 1-B
MAESHIMSSEAPPKTNTGLNFRTNCRMVLNLLREKDCSVTFSPEQPLTPVTDLAVGFSNLSTFSGETPKR
CLDLSNLGDETAPLPTESPDRISSGKVESPKAQFVQFDGLFTPDLGWKAKKCPRGNMNSVLPRLLCSTPS
FKKTSGGQRSVSNKENEGELFKSPNCKPVALLLPQEVVDSQFSPTPENKVDISLDEDCEMNILGSPISAD
PPCLDGAHDDIKMQNLDGFADFFSVDEEEMENPPGAVGNLSSSMAILLSGPLLNQDIEVSNVNNISLNRS
RLYRSPSMPEKLDRPMLKRPVRPLDSETPVRVKRRRSTSSSLQPQEENFQPQRRGTSLKKTLSLCDVDIS
TVLDEDCGHRQLIGDFTKVYALPTVTGRHQDLRYITGETLAALIHGDFSSLVEKIFIIDCRYPYEYDGGH
IKGALNLHRQEEVTDYFLKQPLTPTMAQKRLIIIFHCEFSSERGPKMCRFLREEDRARNEYPSLYYPELY
LLKGGYKDFFPEYKELCEPQSYCPMHHQDFREELLKFRTKCKTSVGDRKRREQIARIMKL
Show Domain Info by CDD
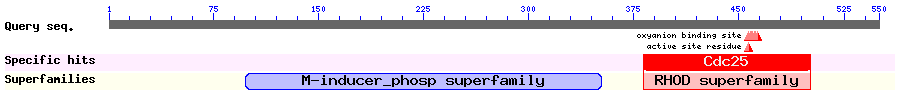
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 988899998999999889755334432133025578987569999998122210002345
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHCCCCCC
AA: MAESHIMSSEAPPKTNTGLNFRTNCRMVLNLLREKDCSVTFSPEQPLTPVTDLAVGFSNL
10 20 30 40 50 60
Conf: 788899986447898987899999999999999999899842223334765675342122
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: STFSGETPKRCLDLSNLGDETAPLPTESPDRISSGKVESPKAQFVQFDGLFTPDLGWKAK
70 80 90 100 110 120
Conf: 499999888898777889998899999989998756787568999998744686322588
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: KCPRGNMNSVLPRLLCSTPSFKKTSGGQRSVSNKENEGELFKSPNCKPVALLLPQEVVDS
130 140 150 160 170 180
Conf: 899999985568799821112589999899999989988432345678768767410001
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHC
AA: QFSPTPENKVDISLDEDCEMNILGSPISADPPCLDGAHDDIKMQNLDGFADFFSVDEEEM
190 200 210 220 230 240
Conf: 699998899994034420389566542346778888887757789999988899998898
Pred: CCCCCCCCCCCCHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: ENPPGAVGNLSSSMAILLSGPLLNQDIEVSNVNNISLNRSRLYRSPSMPEKLDRPMLKRP
250 260 270 280 290 300
Conf: 999999993233455678999898644578866688752236899443100002467987
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCC
AA: VRPLDSETPVRVKRRRSTSSSLQPQEENFQPQRRGTSLKKTLSLCDVDISTVLDEDCGHR
310 320 330 340 350 360
Conf: 776777664224644688999876699999999816543440408999504765325985
Pred: CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHCCCCCCCCCCEEEEEECCCCCCCCCC
AA: QLIGDFTKVYALPTVTGRHQDLRYITGETLAALIHGDFSSLVEKIFIIDCRYPYEYDGGH
370 380 390 400 410 420
Conf: 577201599989999962499998989997799994523788203110056785414999
Pred: CCCCCCCCCHHHHHHHHHCCCCCCCCCCCCEEEEEECCCCCCCCCHHHHHHHHHHHCCCC
AA: IKGALNLHRQEEVTDYFLKQPLTPTMAQKRLIIIFHCEFSSERGPKMCRFLREEDRARNE
430 440 450 460 470 480
Conf: 999999827984588300222134899999962389531399999999751033321132
Pred: CCCCCCCCEEEECCCCCCHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHCCCCCCHHH
AA: YPSLYYPELYLLKGGYKDFFPEYKELCEPQSYCPMHHQDFREELLKFRTKCKTSVGDRKR
490 500 510 520 530 540
Conf: 4667641039
Pred: HHHHHHHCCC
AA: REQIARIMKL
550
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 E -1.000 *
4 S -1.000 *
5 H -1.000 *
6 I -1.000 *
7 M -1.000 *
8 S -1.000 *
9 S -1.000 *
10 E -1.000 *
11 A -1.000 *
12 P -1.000 *
13 P -1.000 *
14 K -1.000 *
15 T -1.000 *
16 N -1.000 *
17 T -1.000 *
18 G -1.000 *
19 L -1.000 *
20 N -1.000 *
21 F -1.000 *
22 R -1.000 *
23 T -1.000 *
24 N -1.000 *
25 C -1.000 *
26 R -1.000 *
27 M -1.000 *
28 V -1.000 *
29 L -1.000 *
30 N -1.000 *
31 L -1.000 *
32 L -1.000 *
33 R -1.000 *
34 E -1.000 *
35 K -1.000 *
36 D -1.000 *
37 C -1.000 *
38 S -1.000 *
39 V -1.000 *
40 T -1.000 *
41 F -1.000 *
42 S -1.000 *
43 P -1.000 *
44 E -1.000 *
45 Q -1.000 *
46 P -1.000 *
47 L -1.000 *
48 T -1.000 *
49 P -1.000 *
50 V -1.000 *
51 T -1.000 *
52 D -1.000 *
53 L -1.000 *
54 A -1.000 *
55 V -1.000 *
56 G -1.000 *
57 F -1.000 *
58 S -1.000 *
59 N -1.000 *
60 L -1.000 *
61 S -1.000 *
62 T -1.000 *
63 F -1.000 *
64 S -1.000 *
65 G -1.000 *
66 E -1.000 *
67 T -1.000 *
68 P -1.000 *
69 K -1.000 *
70 R -1.000 *
71 C -1.000 *
72 L -1.000 *
73 D -1.000 *
74 L -1.000 *
75 S -1.000 *
76 N -1.000 *
77 L -1.000 *
78 G -1.000 *
79 D -1.000 *
80 E -1.000 *
81 T -1.000 *
82 A -1.000 *
83 P -1.000 *
84 L -0.499
85 P -0.420
86 T 0.009
87 E -0.177
88 S 0.470
89 P 0.079
90 D 0.059
91 R -0.371
92 I -0.153
93 S -0.390
94 S -0.302
95 G -0.488
96 K -0.438
97 V 0.107
98 E 0.066
99 S -0.096
100 P -0.028
101 K -0.492
102 A -0.222
103 Q -0.324
104 F -0.464
105 V -1.000 *
106 Q -1.000 *
107 F -1.000 *
108 D -1.000 *
109 G -1.000 *
110 L -1.000 *
111 F -1.000 *
112 T -1.000 *
113 P -1.000 *
114 D -1.000 *
115 L -1.000 *
116 G -1.000 *
117 W -1.000 *
118 K -1.000 *
119 A -1.000 *
120 K -1.000 *
121 K -1.000 *
122 C -1.000 *
123 P -1.000 *
124 R -0.328
125 G -1.000 *
126 N -0.481
127 M -0.505
128 N -0.537
129 S -0.266
130 V -0.497
131 L 0.301
132 P -0.309
133 R 0.108
134 L -0.244
135 L 0.012
136 C -0.163
137 S -0.185
138 T 0.530
139 P 1.187
140 S -0.385
141 F -0.469
142 K -0.398
143 K -0.498
144 T -0.560
145 S -0.498
146 G -0.506
147 G -0.707
148 Q -0.723
149 R -0.490
150 S -0.497
151 V -0.626
152 S -0.532
153 N -0.685
154 K -0.514
155 E -0.428
156 N -0.441
157 E -0.449
158 G -0.467
159 E -0.681
160 L -0.723
161 F -0.552
162 K -0.477
163 S -0.543
164 P -0.189
165 N -0.748
166 C -0.742
167 K -0.669
168 P -0.288
169 V -0.562
170 A -0.594
171 L -0.693
172 L -0.648
173 L -0.480
174 P -0.581
175 Q -0.693
176 E -0.560
177 V -0.613
178 V -0.586
179 D -0.451
180 S -0.449
181 Q -0.621
182 F -0.667
183 S -0.584
184 P -0.414
185 T -0.216
186 P -0.350
187 E -0.607
188 N -0.464
189 K -0.580
190 V -0.521
191 D -0.685
192 I -0.617
193 S -0.655
194 L -0.640
195 D -0.740
196 E -0.498
197 D -0.603
198 C -0.688
199 E -0.397
200 M -0.629
201 N -0.644
202 I -0.668
203 L -0.727
204 G -0.599
205 S -0.504
206 P -0.349
207 I -0.739
208 S -0.608
209 A -0.515
210 D -0.574
211 P -0.672
212 P -0.747
213 C -0.769
214 L -0.652
215 D -0.686
216 G -0.540
217 A -0.646
218 H -0.661
219 D -0.678
220 D -0.653
221 I -0.606
222 K -0.686
223 M -0.644
224 Q -0.586
225 N -0.507
226 L -0.596
227 D -0.527
228 G -0.637
229 F -0.721
230 A -0.624
231 D -0.646
232 F -0.500
233 F -0.722
234 S -0.552
235 V -0.635
236 D -0.414
237 E -0.662
238 E -0.512
239 E -0.544
240 M -0.633
241 E -0.570
242 N -0.711
243 P -0.449
244 P -0.658
245 G -0.640
246 A -0.642
247 V -0.615
248 G -0.677
249 N -0.449
250 L -0.673
251 S -0.550
252 S -0.650
253 S -0.544
254 M -0.589
255 A -0.587
256 I -0.723
257 L -0.652
258 L -0.677
259 S -0.578
260 G -0.394
261 P -0.170
262 L -0.368
263 L -0.661
264 N -0.602
265 Q -0.726
266 D -0.612
267 I -0.757
268 E -0.634
269 V -0.709
270 S -0.495
271 N -0.535
272 V -0.718
273 N -0.591
274 N -0.550
275 I -0.601
276 S -0.735
277 L -0.808
278 N -0.579
279 R -0.320
280 S -0.725
281 R -0.512
282 L -0.554
283 Y -0.676
284 R -0.270
285 S -0.275
286 P -0.232
287 S -0.305
288 M -0.539
289 P -0.554
290 E -0.568
291 K -0.593
292 L -0.692
293 D -0.537
294 R -0.188
295 P -0.163
296 M -0.580
297 L -0.673
298 K -0.485
299 R -0.481
300 P -0.592
301 V -0.494
302 R -0.526
303 P -0.732
304 L -0.764
305 D -0.337
306 S -0.627
307 E -0.560
308 T -0.398
309 P -0.486
310 V -0.593
311 R -0.585
312 V -0.686
313 K -0.468
314 R -0.574
315 R -0.548
316 R -0.534
317 S -0.614
318 T -0.631
319 S -0.585
320 S -0.636
321 S -0.580
322 L -0.720
323 Q -0.717
324 P -0.568
325 Q -0.681
326 E -0.622
327 E -0.648
328 N -0.719
329 F -0.747
330 Q -0.565
331 P -0.527
332 Q -0.618
333 R -0.502
334 R -0.651
335 G -0.685
336 T -0.683
337 S -0.749
338 L -0.417
339 K -0.502
340 K -0.286
341 T -0.440
342 L -0.554
343 S 0.143
344 L -0.390
345 C -0.703
346 D -0.493
347 V -0.538
348 D -0.410
349 I -0.413
350 S -0.587
351 T -0.633
352 V -0.168
353 L -0.269
354 D -0.474
355 E -0.475
356 D -0.204
357 C -0.724
358 G -0.575
359 H -0.489
360 R -0.525
361 Q -0.604
362 L -0.323
363 I -0.070
364 G -0.374
365 D 0.278
366 F -0.432
367 T -0.406
368 K -0.151
369 V -0.540
370 Y -0.168
371 A -0.402
372 L 1.092
373 P 0.454
374 T 0.019
375 V -0.188
376 T -0.569
377 G -0.079
378 R -0.159
379 H -0.028
380 Q -0.220
381 D -0.033
382 L 0.769
383 R 0.428
384 Y 0.317
385 I 2.606
386 T 0.399
387 G -0.056
388 E -0.123
389 T 0.607
390 L 1.246
391 A -0.356
392 A -0.040
393 L 0.830
394 I 1.182
395 H -0.268
396 G 0.640
397 D -0.359
398 F 0.345
399 S -0.205
400 S -0.178
401 L -0.637
402 V 0.210
403 E -0.005
404 K -0.013
405 I 0.031
406 F -0.224
407 I 1.637
408 I 1.960
409 D 3.224
410 C 2.908
411 R 3.224
412 Y 2.431
413 P 0.767
414 Y 2.267
415 E 3.794
416 Y 2.713
417 D -0.123
418 G 2.099
419 G 3.224
420 H 2.342
421 I 2.003
422 K -0.283
423 G 0.510
424 A 3.106
425 L 0.419
426 N 2.448
427 L 0.579
428 H -0.062
429 R -0.083
430 Q -0.032
431 E -0.006
432 E -0.442
433 V 1.001
434 T -0.485
435 D -0.211
436 Y -0.492
437 F 1.506
438 L 0.180
439 K -0.290
440 Q -0.485
441 P -0.028
442 L -0.661
443 T -0.592
444 P -0.432
445 T -0.622
446 M -0.604
447 A -0.544
448 Q -0.553
449 K -0.257
450 R 0.272
451 L -0.352
452 I 0.804
453 I 1.078
454 I 1.999
455 F 2.118
456 H 2.030
457 C 3.249
458 E 3.249
459 F 2.095
460 S 3.210
461 S 0.225
462 E 0.006
463 R 2.772
464 G 1.940
465 P 3.402
466 K -0.342
467 M 0.321
468 C 0.764
469 R 0.215
470 F -0.026
471 L 1.288
472 R 3.416
473 E -0.041
474 E -0.525
475 D 3.416
476 R 3.057
477 A -0.575
478 R -0.530
479 N 2.236
480 E -0.174
481 Y 1.768
482 P 2.967
483 S -0.448
484 L 1.306
485 Y -0.005
486 Y 2.195
487 P 2.227
488 E 1.622
489 L 1.162
490 Y 1.771
491 L 1.890
492 L 2.375
493 K 0.283
494 G 0.735
495 G 3.794
496 Y 2.829
497 K 0.695
498 D 0.132
499 F 2.040
500 F 1.895
501 P -0.413
502 E -0.302
503 Y 0.031
504 K -0.228
505 E -0.449
506 L 0.031
507 C 2.440
508 E -0.058
509 P 2.715
510 Q -0.382
511 S -0.034
512 Y 2.992
513 C 0.547
514 P 0.099
515 M 3.794
516 H -0.327
517 H 0.997
518 Q -0.081
519 D 0.010
520 F 1.381
521 R -0.136
522 E -0.186
523 E 0.536
524 L 1.139
525 L -0.366
526 K -0.367
527 F 0.282
528 R 0.314
529 T -0.511
530 K 0.004
531 C -0.093
532 K 0.607
533 T -0.033
534 S 0.077
535 V -0.340
536 G -0.061
537 D 0.092
538 R -0.170
539 K 0.130
540 R 0.044
541 R 0.160
542 E -0.289
543 Q -0.283
544 I 0.305
545 A -0.214
546 R 0.835
547 I 0.855
548 M 0.221
549 K 1.053
550 L -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 46.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MAESHIMSSE APPKTNTGLN FRTNCRMVLN LLREKDCSVT FSPEQPLTPV TDLAVGFSNL
70 80 90 100 110 120
STFSGETPKR CLDLSNLGDE TAPLPTESPD RISSGKVESP KAQFVQFDGL FTPDLGWKAK
130 140 150 160 170 180
KCPRGNMNSV LPRLLCSTPS FKKTSGGQRS VSNKENEGEL FKSPNCKPVA LLLPQEVVDS
190 200 210 220 230 240
QFSPTPENKV DISLDEDCEM NILGSPISAD PPCLDGAHDD IKMQNLDGFA DFFSVDEEEM
250 260 270 280 290 300
ENPPGAVGNL SSSMAILLSG PLLNQDIEVS NVNNISLNRS RLYRSPSMPE KLDRPMLKRP
310 320 330 340 350 360
VRPLDSETPV RVKRRRSTSS SLQPQEENFQ PQRRGTSLKK TLSLCDVDIS TVLDEDCGHR
370 380 390 400 410 420
QLIGDFTKVY ALPTVTGRHQ DLRYITGETL AALIHGDFSS LVEKIFIIDC RYPYEYDGGH
430 440 450 460 470 480
IKGALNLHRQ EEVTDYFLKQ PLTPTMAQKR LIIIFHCEFS SERGPKMCRF LREEDRARNE
490 500 510 520 530 540
YPSLYYPELY LLKGGYKDFF PEYKELCEPQ SYCPMHHQDF REELLKFRTK CKTSVGDRKR
550
REQIARIMKL
3D Structures in PDB
Not Available
Comments
Cdc25 (Cdc25C) activates the protein kinase Cdc2, a maturation-promoting factor during mitosis, by dephosphorylates T14 and Y15. The localization of Cdc25 is regulated in part by 14-3-3 protein. Kumagai et al. found that When expressed at low level, Cdc25 is mainly cytoplasmic. A mutant form of Cdc25 with S287/A, which cannot bind Cdc25 is almost exclusively nuclear. Mutation of a putative bipartite NLS (aa 298-316) makes the protein cytoplasmic, indicating that aa 298-316 is essential for its nuclear entry. The binding of 14-3-3 near the NLS, likely masking the NLS, prevents its nuclear localization. They also found that LMB treatment elicited a rapid redistribution of Cdc25 from cytoplasm to the nucleus, indicating that the protein is actively exported by CRM1. Mutations of either L53A/V55A/F57A or L344A/V347A/I349A make the protein nuclear. Mutations of potential NES in the protein 14-3-3 did not show effect on localization of both proteins and the wild type 14-3-3 cannot promote the export of mutated Cdc25 indicating that the NES of Cdc25 is responsible for its export.
References
[1]. "Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25"
Kumagai, A., Dunphy, W.G. (1999)
Genes Dev,
13:1067-1072
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
