Show FASTA Format
>gi|158939322|sp|P46108.2|CRK_HUMAN RecName: Full=Adapter molecule crk; AltName: Full=Proto-oncogene c-Crk; AltName: Full=p38
MAGNFDSEERSSWYWGRLSRQEAVALLQGQRHGVFLVRDSSTSPGDYVLSVSENSRVSHYIINSSGPRPP
VPPSPAQPPPGVSPSRLRIGDQEFDSLPALLEFYKIHYLDTTTLIEPVSRSRQGSGVILRQEEAEYVRAL
FDFNGNDEEDLPFKKGDILRIRDKPEEQWWNAEDSEGKRGMIPVPYVEKYRPASASVSALIGGNQEGSHP
QPLGGPEPGPYAQPSVNTPLPNLQNGPIYARVIQKRVPNAYDKTALALEVGELVKVTKINVSGQWEGECN
GKRGHFPFTHVRLLDQQNPDEDFS
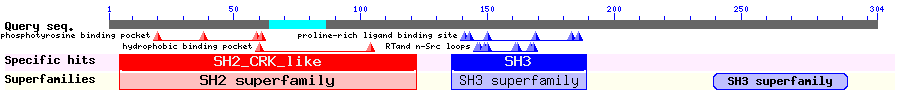
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 988888889998776668999999985699872289762789999629998229934579
Pred: CCCCCCCCCCCCCCCCCCCHHHHHHHHCCCCCCEEEEECCCCCCCCEEEEEECCCCEEEE
AA: MAGNFDSEERSSWYWGRLSRQEAVALLQGQRHGVFLVRDSSTSPGDYVLSVSENSRVSHY
10 20 30 40 50 60
Conf: 985789999999999999999999607742032598457787551044211024304754
Pred: EECCCCCCCCCCCCCCCCCCCCCCCCEEEECCCCCCHHHHHHHHHCCCCCCCEEEECCCC
AA: IINSSGPRPPVPPSPAQPPPGVSPSRLRIGDQEFDSLPALLEFYKIHYLDTTTLIEPVSR
70 80 90 100 110 120
Conf: 568986434335530699940378999999889999679972469998156334999454
Pred: CCCCCCCCCCCCCCCEEEEEECCCCCCCCCCCCCCCCEEEEEECCCCCEEEEECCCCCEE
AA: SRQGSGVILRQEEAEYVRALFDFNGNDEEDLPFKKGDILRIRDKPEEQWWNAEDSEGKRG
130 140 150 160 170 180
Conf: 441897564368983422312799999999999999999888886366799988997258
Pred: EEECCCEEECCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCEE
AA: MIPVPYVEKYRPASASVSALIGGNQEGSHPQPLGGPEPGPYAQPSVNTPLPNLQNGPIYA
190 200 210 220 230 240
Conf: 999823689566555013308859998706997568855995667406754735898999
Pred: EEEECCCCCCCCCCCCEEEECCEEEEEEECCCCEEEEEECCEEEEECCCCEEECCCCCCC
AA: RVIQKRVPNAYDKTALALEVGELVKVTKINVSGQWEGECNGKRGHFPFTHVRLLDQQNPD
250 260 270 280 290 300
Conf: 9999
Pred: CCCC
AA: EDFS
Show Conservation Score by AL2CO
1 M -1.000 *
2 A -1.000 *
3 G -1.000 *
4 N -1.000 *
5 F -1.000 *
6 D 1.569
7 S -0.280
8 E -0.533
9 E -0.598
10 R -0.766
11 S -0.498
12 S -0.223
13 W 3.530
14 Y 2.441
15 W -0.351
16 G 0.293
17 R -0.495
18 L 0.949
19 S 0.215
20 R 3.929
21 Q -0.763
22 E -0.430
23 A 2.104
24 V 0.369
25 A -0.451
26 L -0.033
27 L 2.781
28 Q -0.359
29 G -0.690
30 Q -0.638
31 R -0.371
32 H -0.060
33 G 2.998
34 V 0.148
35 F 2.257
36 L 2.187
37 V 1.549
38 R 3.472
39 D -0.330
40 S 1.670
41 S -0.280
42 T 0.404
43 S -0.671
44 P -0.480
45 G 0.345
46 D 0.219
47 Y 1.399
48 V 1.004
49 L 1.641
50 S 1.864
51 V 1.163
52 S 0.579
53 E -0.338
54 N -0.314
55 S -0.387
56 R -0.047
57 V 0.797
58 S 0.085
59 H 1.869
60 Y 0.701
61 I 1.120
62 I 1.528
63 N -0.367
64 S 0.072
65 S -0.497
66 G -0.640
67 P -1.000 *
68 R -1.000 *
69 P -1.000 *
70 P -1.000 *
71 V -1.000 *
72 P -1.000 *
73 P -1.000 *
74 S -1.000 *
75 P -1.000 *
76 A -1.000 *
77 Q -1.000 *
78 P -1.000 *
79 P -1.000 *
80 P -1.000 *
81 G -1.000 *
82 V -1.000 *
83 S -1.000 *
84 P -1.000 *
85 S -0.473
86 R -0.518
87 L 1.324
88 R -0.651
89 I 0.699
90 G -0.207
91 D -0.306
92 Q -0.418
93 E -0.747
94 F 2.760
95 D -0.682
96 S 0.492
97 L 1.434
98 P -0.688
99 A -0.194
100 L 1.248
101 L 1.351
102 E -0.263
103 F 0.692
104 Y 0.632
105 K -0.275
106 I -0.820
107 H -0.370
108 Y -0.650
109 L 0.316
110 D -0.243
111 T -0.341
112 T -0.797
113 T -0.847
114 L -0.727
115 I -0.661
116 E -0.830
117 P -0.713
118 V -0.770
119 S -0.820
120 R -0.643
121 S -0.904
122 R -0.991
123 Q -0.962
124 G -0.935
125 S -0.695
126 G -0.792
127 V -0.760
128 I -0.806
129 L -0.874
130 R -0.918
131 Q -0.975
132 E -0.920
133 E -0.976
134 A -0.837
135 E -1.047
136 Y -0.853
137 V -0.520
138 R -0.057
139 A 0.684
140 L -0.364
141 F -0.398
142 D 0.134
143 F 0.586
144 N -0.650
145 G 0.011
146 N -0.577
147 D -0.535
148 E -0.513
149 E -0.264
150 D 0.704
151 L 1.580
152 P -0.395
153 F 2.310
154 K -0.616
155 K 0.026
156 G 1.029
157 D 0.724
158 I -0.145
159 L 1.105
160 R -0.575
161 I 0.325
162 R 0.244
163 D -0.454
164 K -0.535
165 P -0.659
166 E -0.087
167 E -0.268
168 Q -0.212
169 W 0.442
170 W 0.953
171 N -0.264
172 A 0.995
173 E 0.002
174 D -0.651
175 S -0.608
176 E -0.756
177 G 0.355
178 K -0.267
179 R -0.396
180 G 0.508
181 M -0.518
182 I 1.190
183 P 0.819
184 V -0.136
185 P -0.082
186 Y 1.269
187 V 0.942
188 E -0.073
189 K -0.582
190 Y -0.507
191 R -0.767
192 P -0.645
193 A -0.901
194 S -0.985
195 A -0.787
196 S -0.922
197 V -0.891
198 S -0.666
199 A -0.815
200 L -0.893
201 I -0.884
202 G -0.804
203 G -0.951
204 N -0.784
205 Q -0.792
206 E -0.705
207 G -0.876
208 S -0.926
209 H -0.833
210 P -0.772
211 Q -0.821
212 P -0.588
213 L -0.787
214 G -0.727
215 G -0.722
216 P -0.981
217 E -0.802
218 P -0.767
219 G -0.984
220 P -0.793
221 Y -0.801
222 A -0.769
223 Q -0.995
224 P -0.800
225 S -0.807
226 V -0.937
227 N -0.978
228 T -0.809
229 P -0.931
230 L -0.813
231 P -0.782
232 N -0.779
233 L -0.783
234 Q -0.640
235 N -0.663
236 G -0.936
237 P -0.370
238 I -0.847
239 Y -0.826
240 A -0.637
241 R -0.761
242 V -0.352
243 I -0.250
244 Q -0.539
245 K -0.507
246 R -0.346
247 V -0.383
248 P -0.280
249 N -0.504
250 A 0.362
251 Y -0.517
252 D 0.312
253 K -0.336
254 T -0.342
255 A 0.883
256 L 0.813
257 A -0.425
258 L 1.504
259 E -0.558
260 V 0.092
261 G 0.709
262 E 0.516
263 L -0.199
264 V 1.274
265 K -0.472
266 V 1.492
267 T 0.029
268 K -0.194
269 I -0.492
270 N -0.334
271 V -0.145
272 S -0.206
273 G 0.722
274 Q 0.501
275 W 1.176
276 E 0.235
277 G 2.720
278 E -0.270
279 C -0.319
280 N -0.254
281 G 0.386
282 K 0.328
283 R -0.129
284 G 1.445
285 H 0.132
286 F 2.716
287 P 4.305
288 F 0.056
289 T 0.752
290 H 1.861
291 V 2.046
292 R 0.533
293 L -1.000 *
294 L -1.000 *
295 D -1.000 *
296 Q -1.000 *
297 Q -1.000 *
298 N -1.000 *
299 P -1.000 *
300 D -1.000 *
301 E -1.000 *
302 D -1.000 *
303 F -1.000 *
304 S -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 57.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
MAGNFDSEER SSWYWGRLSR QEAVALLQGQ RHGVFLVRDS STSPGDYVLS VSENSRVSHY
70 80 90 100 110 120
IINSSGPRPP VPPSPAQPPP GVSPSRLRIG DQEFDSLPAL LEFYKIHYLD TTTLIEPVSR
130 140 150 160 170 180
SRQGSGVILR QEEAEYVRAL FDFNGNDEED LPFKKGDILR IRDKPEEQWW NAEDSEGKRG
190 200 210 220 230 240
MIPVPYVEKY RPASASVSAL IGGNQEGSHP QPLGGPEPGP YAQPSVNTPL PNLQNGPIYA
250 260 270 280 290 300
RVIQKRVPNA YDKTALALEV GELVKVTKIN VSGQWEGECN GKRGHFPFTH VRLLDQQNPD
EDFS
Crk is a small adapter protein with one SH2 domain and two SH3 domains. Through binding of certain proteins to SH2 and SH3 domain, Crk is implicated in several signaling processes. In particular, binding of phosphorylated Wee1 to SH2 domain is required for the apoptosis. In the second SH3 domain of Crk, there is an NES interacting with Crm1. Removal of the NES enhances Wee1-Crk binding and increases the proapoptotic activity of the complex.
[1]. "Apoptotic regulation by the Crk adapter protein mediated by interactions with Wee1 and Crm1/exportin."
Smith JJ, Richardson DA, Kopf J, Yoshida M, Hollingsworth RE, Kornbluth S (2002)
Mol Cell Biol,
22:1412-23
PubMed
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
