Summary for Mst1 (NES ID: 68)
Full Name
Serine/threonine-protein kinase 4
UniProt Alternative Names
STE20-like kinase MST1
Mammalian STE20-like protein kinase 1
Serine/threonine-protein kinase Krs-2
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
Mutations That Affect Nuclear Export
Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
*shown as underlined residues in the full sequence
323DEMDSGTMVRAVGDEMGTVRVASTMTDGANTMIEHDDTLPSQLGTMVINAEDEEEEGTM
381,
430DGDYEFLKSWTVEDLQKRLLALDPMMEQEIE
460
Secondary Structure of Export Signal
Unknown
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
>gi|13124559|sp|Q13043.2|STK4_HUMAN RecName: Full=Serine/threonine-protein kinase 4; AltName: Full=Mammalian STE20-like protein kinase 1; Short=MST-1; AltName: Full=STE20-like kinase MST1; AltName: Full=Serine/threonine-protein kinase Krs-2
METVQLRNPPRRQLKKLDEDSLTKQPEEVFDVLEKLGEGSYGSVYKAIHKETGQIVAIKQVPVESDLQEI
IKEISIMQQCDSPHVVKYYGSYFKNTDLWIVMEYCGAGSVSDIIRLRNKTLTEDEIATILQSTLKGLEYL
HFMRKIHRDIKAGNILLNTEGHAKLADFGVAGQLTDTMAKRNTVIGTPFWMAPEVIQEIGYNCVADIWSL
GITAIEMAEGKPPYADIHPMRAIFMIPTNPPPTFRKPELWSDNFTDFVKQCLVKSPEQRATATQLLQHPF
VRSAKGVSILRDLINEAMDVKLKRQESQQREVDQDDEENSEEDEMDSGTMVRAVGDEMGTVRVASTMTDG
ANTMIEHDDTLPSQLGTMVINAEDEEEEGTMKRRDETMQPAKPSFLEYFEQKEKENQINSFGKSVPGPLK
NSSDWKIPQDGDYEFLKSWTVEDLQKRLLALDPMMEQEIEEIRQKYQSKRQPILDAIEAKKRRQQNF
Show Domain Info by CDD
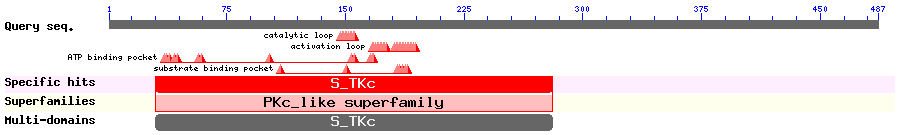
Show Secondary Structure by PSIPRED
# PSIPRED HFORMAT (PSIPRED V3.2)
Conf: 941235699800002379320268924788987763677751134655536897899997
Pred: CCCCCCCCCCCHHCCCCCCCCCCCCCHHHHHHHHHHCCCCCCEEEEEEECCCCCEEEEEE
AA: METVQLRNPPRRQLKKLDEDSLTKQPEEVFDVLEKLGEGSYGSVYKAIHKETGQIVAIKQ
10 20 30 40 50 60
Conf: 058750799999998773189996101421076187069998504987388988741999
Pred: CCCCCHHHHHHHHHHHHHHCCCCCCCEEEEEEEECCEEEEEEEECCCCCHHHHHHHCCCC
AA: VPVESDLQEIIKEISIMQQCDSPHVVKYYGSYFKNTDLWIVMEYCGAGSVSDIIRLRNKT
70 80 90 100 110 120
Conf: 887899999998642025331498410001356310068896343110134454664311
Pred: CCHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCCCEEECCCCCEEEECCHHHHHHHHHHHC
AA: LTEDEIATILQSTLKGLEYLHFMRKIHRDIKAGNILLNTEGHAKLADFGVAGQLTDTMAK
130 140 150 160 170 180
Conf: 463112565552788866246542322210256987305999988988331110146999
Pred: CCCCCCCCCCCCHHHHHHHCCCCCCCEECCHHHHHHHCCCCCCCCCCCCCEEEECCCCCC
AA: RNTVIGTPFWMAPEVIQEIGYNCVADIWSLGITAIEMAEGKPPYADIHPMRAIFMIPTNP
190 200 210 220 230 240
Conf: 998899988894589999874044999976877651593201577700489999999999
Pred: CCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCHHHHHCCCCCCCCCCCHHHHHHHHHHHHH
AA: PPTFRKPELWSDNFTDFVKQCLVKSPEQRATATQLLQHPFVRSAKGVSILRDLINEAMDV
250 260 270 280 290 300
Conf: 987333332101223455775553357841003789887300034457999941005998
Pred: HHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
AA: KLKRQESQQREVDQDDEENSEEDEMDSGTMVRAVGDEMGTVRVASTMTDGANTMIEHDDT
310 320 330 340 350 360
Conf: 989875123338997755653445887899999813444555431027999988999899
Pred: CCCCCCCEEECCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHCCCCCCCCCCCCCC
AA: LPSQLGTMVINAEDEEEEGTMKRRDETMQPAKPSFLEYFEQKEKENQINSFGKSVPGPLK
370 380 390 400 410 420
Conf: 999999999999544688896788986303992679999999985203650479999999
Pred: CCCCCCCCCCCCCCCCCCCCHHHHHHHHHCCCHHHHHHHHHHHHHHCCCCCHHHHHHHHH
AA: NSSDWKIPQDGDYEFLKSWTVEDLQKRLLALDPMMEQEIEEIRQKYQSKRQPILDAIEAK
430 440 450 460 470 480
Conf: 9973069
Pred: HHHHHCC
AA: KRRQQNF
Show Conservation Score by AL2CO
1 M -1.000 *
2 E -1.000 *
3 T -1.000 *
4 V -1.000 *
5 Q -1.000 *
6 L -1.000 *
7 R -1.000 *
8 N -1.000 *
9 P -1.000 *
10 P -1.000 *
11 R -1.000 *
12 R -1.000 *
13 Q -1.000 *
14 L -1.000 *
15 K -1.000 *
16 K -1.000 *
17 L -1.000 *
18 D -1.000 *
19 E -1.000 *
20 D -1.000 *
21 S -0.461
22 L -0.847
23 T -0.837
24 K -0.693
25 Q -0.425
26 P 0.662
27 E -0.657
28 E -0.731
29 V -0.841
30 F 0.856
31 D -0.539
32 V -0.529
33 L -0.498
34 E -0.741
35 K -0.244
36 L 0.883
37 G 2.277
38 E -0.335
39 G 2.277
40 S 0.680
41 Y 0.928
42 G 1.610
43 S -0.670
44 V 2.277
45 Y 0.526
46 K 0.246
47 A 0.876
48 I -0.648
49 H -0.328
50 K -0.406
51 E -0.801
52 T 0.147
53 G -0.440
54 Q -0.727
55 I -0.766
56 V 0.214
57 A 1.612
58 I 0.741
59 K 1.921
60 Q -0.443
61 V 0.523
62 P -0.240
63 V 0.234
64 E 0.049
65 S -0.259
66 D 0.247
67 L -0.284
68 Q -0.494
69 E 0.022
70 I 0.343
71 I -0.262
72 K -0.358
73 E 1.918
74 I 1.004
75 S -0.735
76 I -0.073
77 M 0.951
78 Q -0.089
79 Q -0.208
80 C 0.089
81 D -0.569
82 S 0.108
83 P -0.696
84 H 0.097
85 V 0.966
86 V 0.701
87 K -0.341
88 Y 0.854
89 Y -0.493
90 G 0.163
91 S 0.756
92 Y 0.546
93 F -0.529
94 K -0.769
95 N -0.505
96 T -0.758
97 D -0.552
98 L 0.895
99 W 0.456
100 I 1.209
101 V 0.352
102 M 1.253
103 E 2.277
104 Y 1.054
105 C 0.906
106 G 0.046
107 A 0.543
108 G 1.911
109 S 1.453
110 V 0.188
111 S -0.432
112 D 0.992
113 I 0.285
114 I 0.024
115 R -0.427
116 L -0.430
117 R -1.000 *
118 N -0.510
119 K -0.862
120 T -0.776
121 L 0.444
122 T -0.786
123 E 1.612
124 D -1.020
125 E -0.589
126 I 0.629
127 A -0.161
128 T -0.323
129 I 0.641
130 L 0.045
131 Q -0.408
132 S -0.180
133 T -0.013
134 L 0.701
135 K -0.613
136 G 1.415
137 L 1.071
138 E -0.850
139 Y 1.235
140 L 1.618
141 H 1.921
142 F -0.872
143 M -0.784
144 R -0.448
145 K 0.240
146 I 0.703
147 H 1.922
148 R 1.922
149 D 2.277
150 I 1.562
151 K 2.277
152 A 0.648
153 G 0.557
154 N 2.277
155 I 1.173
156 L 1.915
157 L 0.810
158 N 0.003
159 T -0.693
160 E -0.847
161 G 1.721
162 H -0.828
163 A 0.440
164 K 1.916
165 L 1.295
166 A 0.766
167 D 2.277
168 F 1.917
169 G 2.277
170 V 1.288
171 A 0.845
172 G 0.665
173 Q 0.345
174 L 1.085
175 T -0.085
176 D -0.598
177 T -0.396
178 M -0.630
179 A -0.573
180 K 0.129
181 R 0.823
182 N -0.640
183 T 1.583
184 V 0.702
185 I 1.267
186 G 2.277
187 T 1.687
188 P 2.277
189 F 0.971
190 W 1.496
191 M 1.860
192 A 1.384
193 P 2.277
194 E 1.917
195 V 1.132
196 I 0.980
197 Q -0.085
198 E -0.425
199 I -0.685
200 G -0.518
201 Y 1.690
202 N 0.553
203 C -0.840
204 V -0.139
205 A 0.614
206 D 2.277
207 I 0.819
208 W 2.277
209 S 1.559
210 L 0.228
211 G 2.277
212 I 1.617
213 T 1.175
214 A 0.507
215 I 0.766
216 E 2.277
217 M 0.963
218 A 0.486
219 E -0.412
220 G -0.110
221 K -0.735
222 P 2.277
223 P 2.277
224 Y 0.051
225 A -0.151
226 D -0.274
227 I -0.424
228 H 0.448
229 P 1.261
230 M 0.462
231 R 1.246
232 A 1.270
233 I 0.968
234 F 0.740
235 M -0.277
236 I 1.657
237 P 0.517
238 T -0.238
239 N -0.320
240 P -0.578
241 P 0.628
242 P 1.609
243 T -0.588
244 F 0.935
245 R -0.725
246 K -0.553
247 P -0.518
248 E -0.572
249 L -0.723
250 W 0.261
251 S 0.753
252 D -0.677
253 N -0.955
254 F 0.340
255 T -0.578
256 D -0.121
257 F 0.971
258 V 0.368
259 K -0.608
260 Q -0.761
261 C 0.433
262 L 1.359
263 V -0.654
264 K 0.373
265 S -0.376
266 P 0.036
267 E -0.719
268 Q -0.561
269 R 2.277
270 A -0.355
271 T -0.205
272 A 0.468
273 T -0.820
274 Q -0.630
275 L 1.107
276 L 0.512
277 Q -0.641
278 H -0.020
279 P -0.567
280 F 1.184
281 V 0.430
282 R -0.654
283 S -0.825
284 A -0.785
285 K -0.753
286 G -0.518
287 V -0.857
288 S -0.794
289 I -0.977
290 L 0.099
291 R -0.753
292 D -0.642
293 L -0.378
294 I 0.581
295 N -0.693
296 E -0.494
297 A -0.813
298 M -0.948
299 D -0.876
300 V -0.756
301 K -0.980
302 L -1.028
303 K -0.873
304 R -0.922
305 Q -1.046
306 E -0.894
307 S -0.904
308 Q -1.008
309 Q -0.965
310 R -0.993
311 E -1.032
312 V -1.058
313 D -0.863
314 Q -0.902
315 D -0.901
316 D -1.017
317 E -1.012
318 E -0.958
319 N -0.983
320 S -0.954
321 E -1.095
322 E -1.033
323 D -1.000
324 E -1.029
325 M -1.083
326 D -0.952
327 S -1.052
328 G -1.030
329 T -0.871
330 M -1.001
331 V -0.951
332 R -0.912
333 A -1.018
334 V -0.964
335 G -0.988
336 D -0.904
337 E -0.929
338 M -1.063
339 G -0.961
340 T -0.850
341 V -0.898
342 R -0.915
343 V -1.072
344 A -0.992
345 S -0.995
346 T -0.981
347 M -1.031
348 T -1.041
349 D -0.924
350 G -0.917
351 A -1.044
352 N -0.879
353 T -0.963
354 M -0.961
355 I -1.025
356 E -0.975
357 H -1.053
358 D -0.950
359 D -0.921
360 T -0.924
361 L -1.008
362 P -0.870
363 S -0.922
364 Q -0.923
365 L -1.014
366 G -0.649
367 T -0.774
368 M -0.910
369 V -0.912
370 I -1.062
371 N -0.878
372 A -0.909
373 E -1.007
374 D -0.912
375 E -0.928
376 E -1.028
377 E -0.888
378 E -1.087
379 G -0.886
380 T -0.711
381 M -0.896
382 K -0.917
383 R -1.014
384 R -1.019
385 D -0.939
386 E -0.973
387 T -0.945
388 M -1.002
389 Q -1.023
390 P -0.938
391 A -0.945
392 K -0.911
393 P -0.881
394 S -0.968
395 F -1.000 *
396 L -1.000 *
397 E -1.000 *
398 Y -1.000 *
399 F -1.000 *
400 E -1.000 *
401 Q -1.000 *
402 K -1.000 *
403 E -1.000 *
404 K -1.000 *
405 E -1.000 *
406 N -1.000 *
407 Q -1.000 *
408 I -1.000 *
409 N -1.000 *
410 S -1.000 *
411 F -1.000 *
412 G -1.000 *
413 K -1.000 *
414 S -1.000 *
415 V -1.000 *
416 P -1.000 *
417 G -1.000 *
418 P -1.000 *
419 L -1.000 *
420 K -1.000 *
421 N -1.000 *
422 S -1.000 *
423 S -1.000 *
424 D -1.000 *
425 W -1.000 *
426 K -1.000 *
427 I -1.000 *
428 P -1.000 *
429 Q -1.000 *
430 D -1.000 *
431 G -1.000 *
432 D -1.000 *
433 Y -1.000 *
434 E -1.000 *
435 F -1.000 *
436 L -1.000 *
437 K -1.000 *
438 S -1.000 *
439 W -1.000 *
440 T -1.000 *
441 V -1.000 *
442 E -1.000 *
443 D -1.000 *
444 L -1.000 *
445 Q -1.000 *
446 K -1.000 *
447 R -1.000 *
448 L -1.000 *
449 L -1.000 *
450 A -1.000 *
451 L -1.000 *
452 D -1.000 *
453 P -1.000 *
454 M -1.000 *
455 M -1.000 *
456 E -1.000 *
457 Q -1.000 *
458 E -1.000 *
459 I -1.000 *
460 E -1.000 *
461 E -1.000 *
462 I -1.000 *
463 R -1.000 *
464 Q -1.000 *
465 K -1.000 *
466 Y -1.000 *
467 Q -1.000 *
468 S -1.000 *
469 K -1.000 *
470 R -1.000 *
471 Q -1.000 *
472 P -1.000 *
473 I -1.000 *
474 L -1.000 *
475 D -1.000 *
476 A -1.000 *
477 I -1.000 *
478 E -1.000 *
479 A -1.000 *
480 K -1.000 *
481 K -1.000 *
482 R -1.000 *
483 R -1.000 *
484 Q -1.000 *
485 Q -1.000 *
486 N -1.000 *
487 F -1.000 *
* gap fraction no less than 0.50; conservation set to M-S
M: mean; S: standard deviation
al2co - The parameters are:
Input alignment file - 68.paln
Output conservation - STDOUT
Weighting scheme - independent-count based
Conservation calculation method - entropy-based
Window size - 1
Conservation normalized to zero mean and unity variance
Gap fraction to suppress calculation - 0.50
10 20 30 40 50 60
METVQLRNPP RRQLKKLDED SLTKQPEEVF DVLEKLGEGS YGSVYKAIHK ETGQIVAIKQ
70 80 90 100 110 120
VPVESDLQEI IKEISIMQQC DSPHVVKYYG SYFKNTDLWI VMEYCGAGSV SDIIRLRNKT
130 140 150 160 170 180
LTEDEIATIL QSTLKGLEYL HFMRKIHRDI KAGNILLNTE GHAKLADFGV AGQLTDTMAK
190 200 210 220 230 240
RNTVIGTPFW MAPEVIQEIG YNCVADIWSL GITAIEMAEG KPPYADIHPM RAIFMIPTNP
250 260 270 280 290 300
PPTFRKPELW SDNFTDFVKQ CLVKSPEQRA TATQLLQHPF VRSAKGVSIL RDLINEAMDV
310 320 330 340 350 360
KLKRQESQQR EVDQDDEENS EEDEMDSGTM VRAVGDEMGT VRVASTMTDG ANTMIEHDDT
370 380 390 400 410 420
LPSQLGTMVI NAEDEEEEGT MKRRDETMQP AKPSFLEYFE QKEKENQINS FGKSVPGPLK
430 440 450 460 470 480
NSSDWKIPQD GDYEFLKSWT VEDLQKRLLA LDPMMEQEIE EIRQKYQSKR QPILDAIEAK
KRRQQNF
3D Structures in PDB
Not Available
Comments
MST is a member of a subfamily of kinases that share high similarity in the catalytic domain with STE20. The non-catalytic C-terminal domain may function as a negative regulatory domain, the removal of which remarkably increases the kinase activity. MST is cleaved and activated by various apoptotic stimuli. Lee and Yonehara showed that MST is a cytoplasmic protein and rapidly relocalizes to the nucleus after LMB treatment. They fused the putative NES (aa 323-381) and observed that a cytoplamsic localization of the fusion protein, indicating that the region is a functional NES. However, the possibility of another functional NES was raised when simultaneous mutations of all four hydrophobic residues in the putative NES failed to alter the cytoplasmic location of the whole protein. A second NES was identified among aa 430-460. MST became nuclear when the hydrophobic residues within both NES regions were changed to alanines. They also identified a bipartitie NLS region (aa 469-483).
References
[1]. "Phosphorylation and dimerization regulate nucleocytoplasmic shuttling of mammalian STE20-like kinase (MST)"
Lee, K.K., Yonehara, S. (2002)
J Biol Chem,
277:12351-12358
PubMedUser Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.
