Summary for AID (NES ID: 75)
Full Name
Activation-induced cytidine deaminase UniProt
Alternative Names
Cytidine aminohydrolase
Organism
Homo sapiens (Human)
Experimental Evidence for CRM1-mediated Export
LMB Sensitive Ref.1Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase, McBride et al., J Exp. Med., 2004 Ref.3Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks, Brar et al., J Biol Chem, 2004 Ref.4Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1, Ito et al., Proc Natl Acad Sci, 2004 Ref.6Consecutive Interactions With HSP90 and eEF1A Underlie a Functional Maturation and Storage Pathway of AID in the Cytoplasm, Methot et al., J Exp. Med., 2015
Mutations That Affect Nuclear Export
*highlighted yellow in the full sequence
L189A, F193A, L196A, L198A Ref.2The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence, Geisberger et al., Proc Natl Acad Sci, 2009 Ref.3Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks, Brar et al., J Biol Chem, 2004 Mutations That Affect CRM1 Binding
Unknown
Functional Export Signals
*shown as underlined residues in the full sequence
188DLRDAFRTLGL198 Ref.1Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase, McBride et al., J Exp. Med., 2004 Secondary Structure of Export Signal
Other Residues Important for Export
Unknown
Sequence
Show FASTA Format
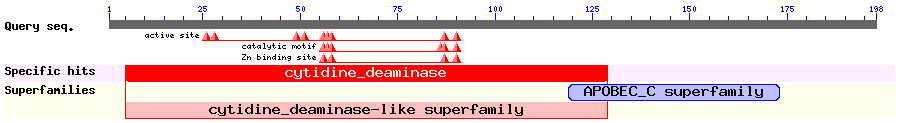
Show Domain Info by CDD
Show Secondary Structure by PSIPRED
Show Conservation Score by AL2CO
10 20 30 40 50 60MDSLLMNRRK FLYQFKNVRW AKGRRETYLC YVVKRRDSAT SFSLDFGYLR NKNGCHVELL70 80 90 100 110 120FLRYISDWDL DPGRCYRVTW FTSWSPCYDC ARHVADFLRG NPNLSLRIFT ARLYFCEDRK130 140 150 160 170 180AEPEGLRRLH RAGVQIAIMT FKDYFYCWNT FVENHERTFK AWEGLHENSV RLSRQLRRIL190LPLYEVDDLR DAFRTLGL
3D Structures in PDB
5W0R (X-Ray,2.4 Å resolution)
Comments
AID is important for antibody gene diversification. It deaminates deoxycytidine to change cytosine to uracil. AID shuttles between the nucleus and the cytoplasm but is mostly cytoplasmic in activated B cells. Removal of C-terminal NES mislocalizes AID to the nucleus and nuclear export of AID is LMB-sensitive. NES sequence in conserved between AID homologues and all are found to have LMB-sensitive nuclear export activity. NES overlaps with region of protein critical for cytoplasmic retention. Computational modeling of the peptide interacting with the catalytic core reveal burying of critical residues for CRM1 interaction, but such conformation may be necessary for interaction with eEF1A and cytoplasmic retention. Ref.6Consecutive Interactions With HSP90 and eEF1A Underlie a Functional Maturation and Storage Pathway of AID in the Cytoplasm, Methot et al., J Exp. Med., 2015
References
[1]. "Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase"
McBride, K.M., Barreto, V., Ramiro, A.R., Stavropoulos, P., Nussenzweig, M.C. (2004) J Exp. Med., 199:1235-1244 PubMed
[2]. "The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence"
Geisberger, R., Rada, C., Neuberger, M.S. (2009) Proc Natl Acad Sci, 106:6736-6741 PubMed
[3]. "Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks"
Brar, S.S., Watson, M., Diaz, M. (2004) J Biol Chem, 279:26395-26401 PubMed
[4]. "Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1"
Ito, S., Nagaoka, H., Shinkura, R., Begum, N., Muramatsu, M., Nakata, M., Honjo, T. (2004) Proc Natl Acad Sci, 101:1975-1980 PubMed
[5]. "Active nuclear import and cytoplasmic retention of activation-induced deaminase"
Patenaude, A.M., Orthwein, A., Hu, Y., Campo, V.A., Kavli, B., Buschiazzo, A., Di Noia, J.M. (2009) Nat. Struct. Mol. Biol., 16:517-527 PubMed
[6]. "Consecutive Interactions With HSP90 and eEF1A Underlie a Functional Maturation and Storage Pathway of AID in the Cytoplasm"
Methot, S.P., Litzler, L.C., Trajtenberg, F., Zahn, A., Robert, F., Pelletier, J., Buschiazzo, A., Magor, B.G., Di Noia, J.M. (2015) J Exp. Med., 212(4):581-96 PubMed
McBride, K.M., Barreto, V., Ramiro, A.R., Stavropoulos, P., Nussenzweig, M.C. (2004) J Exp. Med., 199:1235-1244 PubMed
[2]. "The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence"
Geisberger, R., Rada, C., Neuberger, M.S. (2009) Proc Natl Acad Sci, 106:6736-6741 PubMed
[3]. "Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks"
Brar, S.S., Watson, M., Diaz, M. (2004) J Biol Chem, 279:26395-26401 PubMed
[4]. "Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1"
Ito, S., Nagaoka, H., Shinkura, R., Begum, N., Muramatsu, M., Nakata, M., Honjo, T. (2004) Proc Natl Acad Sci, 101:1975-1980 PubMed
[5]. "Active nuclear import and cytoplasmic retention of activation-induced deaminase"
Patenaude, A.M., Orthwein, A., Hu, Y., Campo, V.A., Kavli, B., Buschiazzo, A., Di Noia, J.M. (2009) Nat. Struct. Mol. Biol., 16:517-527 PubMed
[6]. "Consecutive Interactions With HSP90 and eEF1A Underlie a Functional Maturation and Storage Pathway of AID in the Cytoplasm"
Methot, S.P., Litzler, L.C., Trajtenberg, F., Zahn, A., Robert, F., Pelletier, J., Buschiazzo, A., Magor, B.G., Di Noia, J.M. (2015) J Exp. Med., 212(4):581-96 PubMed
User Input
Accurate identification of NESs is difficult because many sequences in the genome match the NES consensus.
Therefore, some published NESs may be mistakenly identified. Please help us improve the accuracy of NESdb
by providing either a positive or negative flag for the NES in this entry. Supporting comments are required to process the flag.