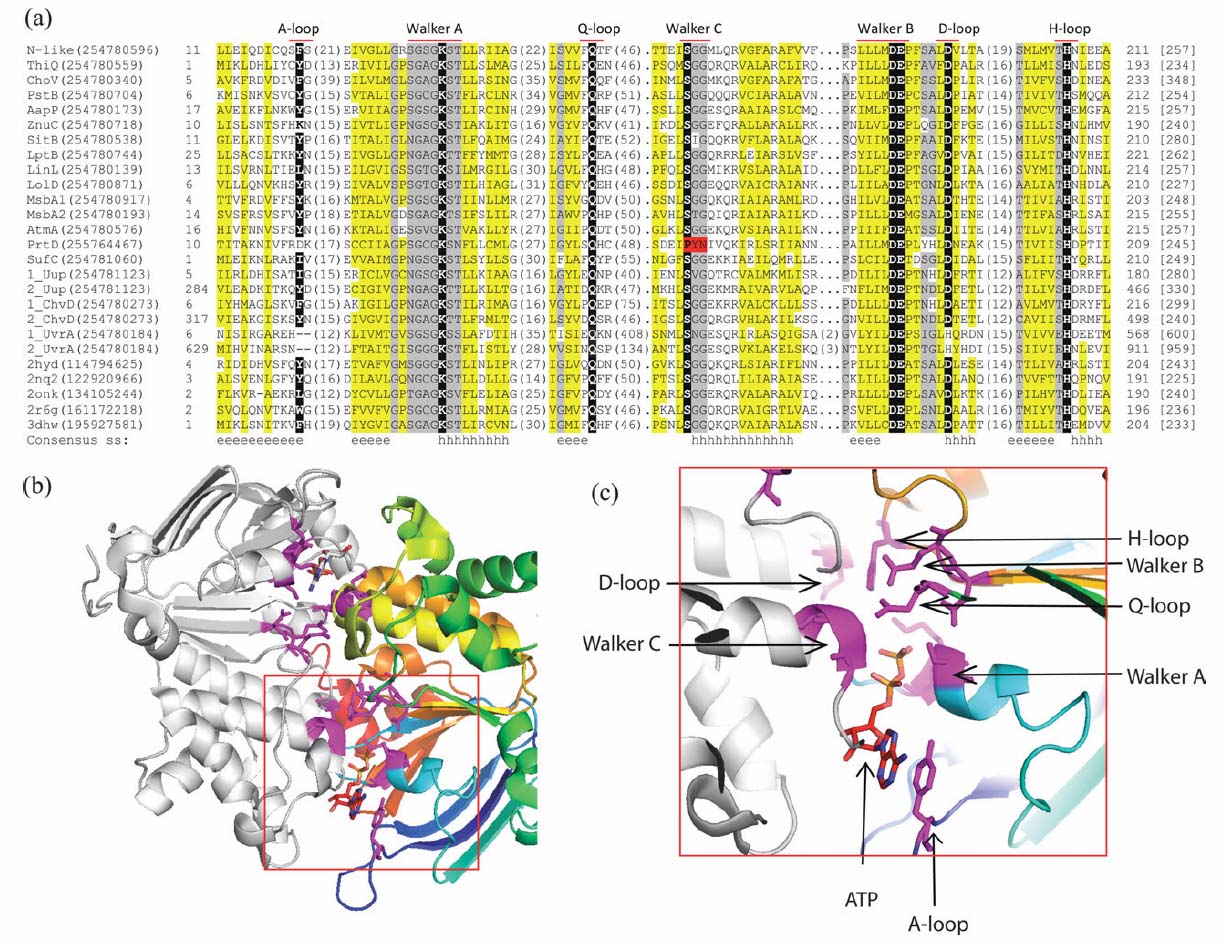
Multiple Sequence Alignment (MSA) and representative structure of Nucleatide Binding Domains (NBDs) in Candidatus Liberibacter asiaticus proteome. (a) Simplified version of the MSA of all NBDs of Candidatus Liberibacter asiaticus and representative homologous structures (only the segments containing sequence motifs of the NBD are shown). The names of motifs are labeled on the top of the MSA. Protein name abbreviations or PDB IDs, with gi number in the parentheses, are used as sequence identifiers at the beginning of each line. N-like is short for Nrt/Ssu/Tau-like system NBD. For ABC-type ATPases with two NBDs, we assign a number in front of the identifier to distinguish between the two domains. In the sequences, hydrophobic residues are highlighted in yellow, small residues positions are colored in gray, and the most essential residues for the function are represented as white letters on black backgrounds. Starting/ending residue numbers and sequence length are shown in italic font and in brackets, respectively. Numbers of residues between the segments are indicated in the parentheses. Dots are used to adjust the space for the MSA. Gaps are shown in dash lines. The PYN residue marked red indicates the substituted Walker C motif. The "consensus ss" line shows the consensus secondary structures predicted by PROMALS3D. For the secondary structure, "e" means beta sheet and "h" stands for alpha helix. (b) Structure of ABC transporter nucleotide-binding domain homodimer with ATP molecules. The structure is adapted from Sav1866 (PDB: 2hyd). The right NBD is colored in rainbow from N to C terminus while the left NBD is colored in gray. Residues essential for the function are shown as sticks in magenta. (c) Close-up of ATP-binding site enlarged from the red frame in (b). ATP and sequence motifs of the NBDs are pointed out.