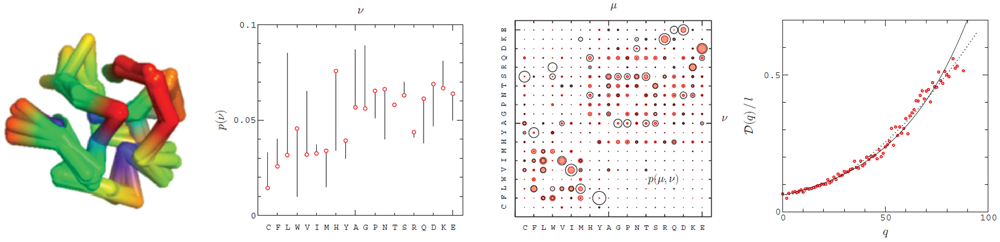
A coarse-grained model recapitulates statistics of protein evolution. (left): Sample of the ensemble, corresponding to the dominant energy basin recovered by a typical viable sequence. The structures are obtained by folding many replicas of the sequence in parallel using methods described in the text. To indicate amino acid type, monomers are colored blue, light blue, blue-green, green, yellow, orange, and red, in order of increasing affinity to solvent. (middle left): Comparison of model and empirical amino acid frequencies, p(ν). Model results are indicated by circles. Differences between model and empirical results are indicated by one-sided error bars. Amino acid labels on the lower axis are arranged in order of increasing affinity to solvent. (middle right): Comparison of model and empirical amino acid replacement probabilities, p(μ, ν). The plot describes the replacement of amino acids of type μ by amino acids of type ν with probabilities indicated by circle radii. Solid red circles indicate model values; open black circles indicate empirical values computed by Dayhoff. (right): Aligned distance between structures, D(q), as a function of the percentage of nonidentical amino acids, q. The solid line is an exponential fit to the data; the dotted line is a power-law fit.