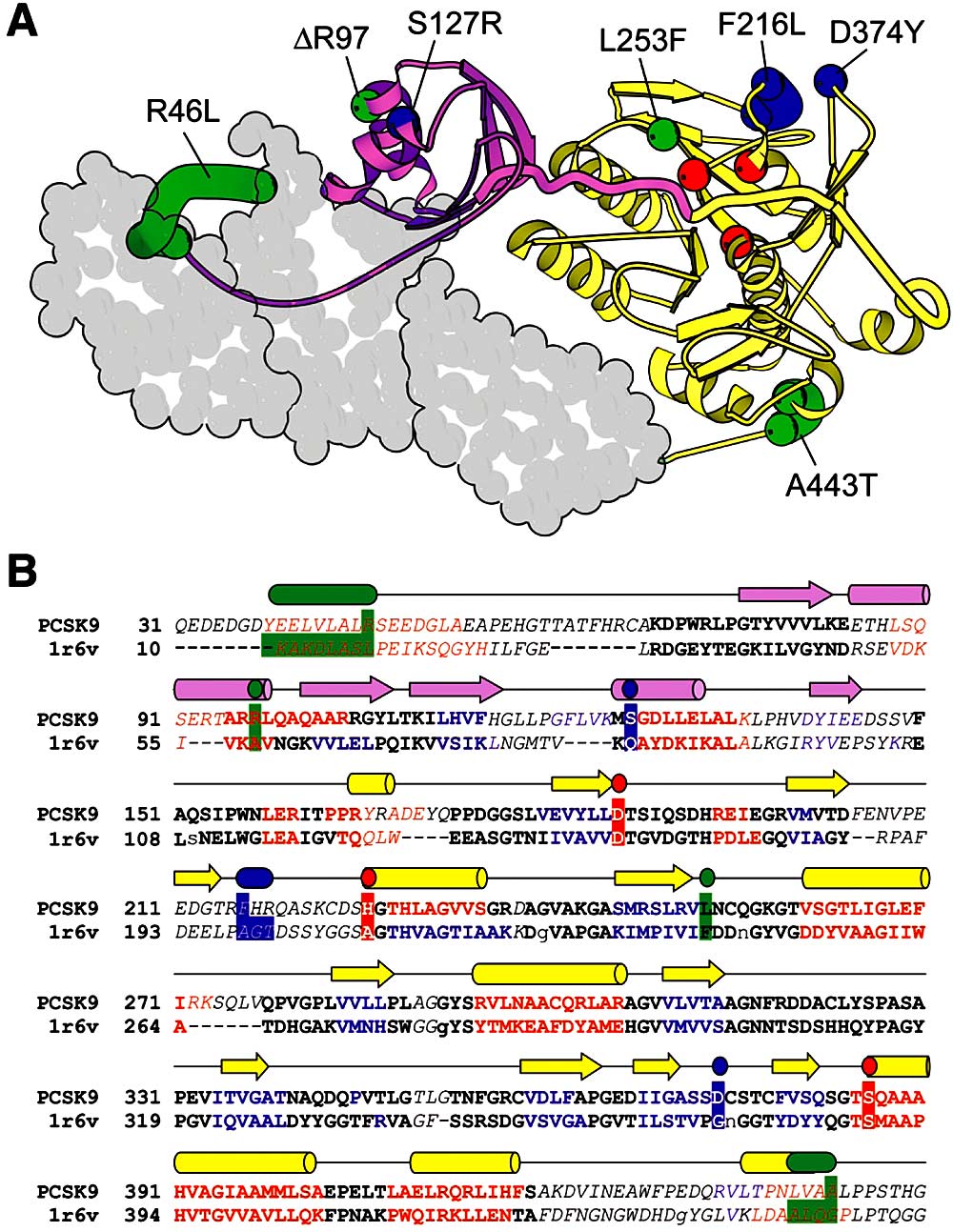
PCSK9 Structure Model. Structural model (A) and alignment of residues 31-450 of PCSK9 with fervidolysin (B). A, A model of PCSK9, generated using coordinates from the crystal structure of fervidolysin (Protein Database ID 1r6v). The N-terminal prodomain (purple) is connected to the catalytic domain (yellow) by a long, thickened coil. The prodomain cleavage site (color boundary) is near the catalytic triad (red). PCSK9 mutations that map to the prodomain or catalytic domain are labeled and numbered according to the human sequence and are depicted in blue (gain of function) or green (loss of function). Those mutations that reside in less accurately mapped regions of the protein are represented by a range of residues. Three predicted C-terminal domains of PCSK9 are represented as a space-fill that is based on the two fervidolysin C-terminal domains. B, An alignment between PCSK9 and the 1r6v N-terminal and catalytic domain sequences, generated with a fold-recognition program (Meta-BASIC). Compared with an alignment made by BLAST (not shown), the residues whose positions in the alignment were in agreement with those obtained using BLAST are in bold type, whereas those that differ are italicized. Noncapitalized residues in 1r6v represent positions where sequence was deleted. Residues with predicted secondary structure are red (helix), blue (strand), and black (coil) and are compared with the observed 1r6v secondary structure (cylinders and arrows shown above the alignment and colored as in panel A). Residues corresponding to the catalytic triad, the gain-of-function mutations, and the loss-of-function mutations are highlighted with the colors and ranges described for panel A.