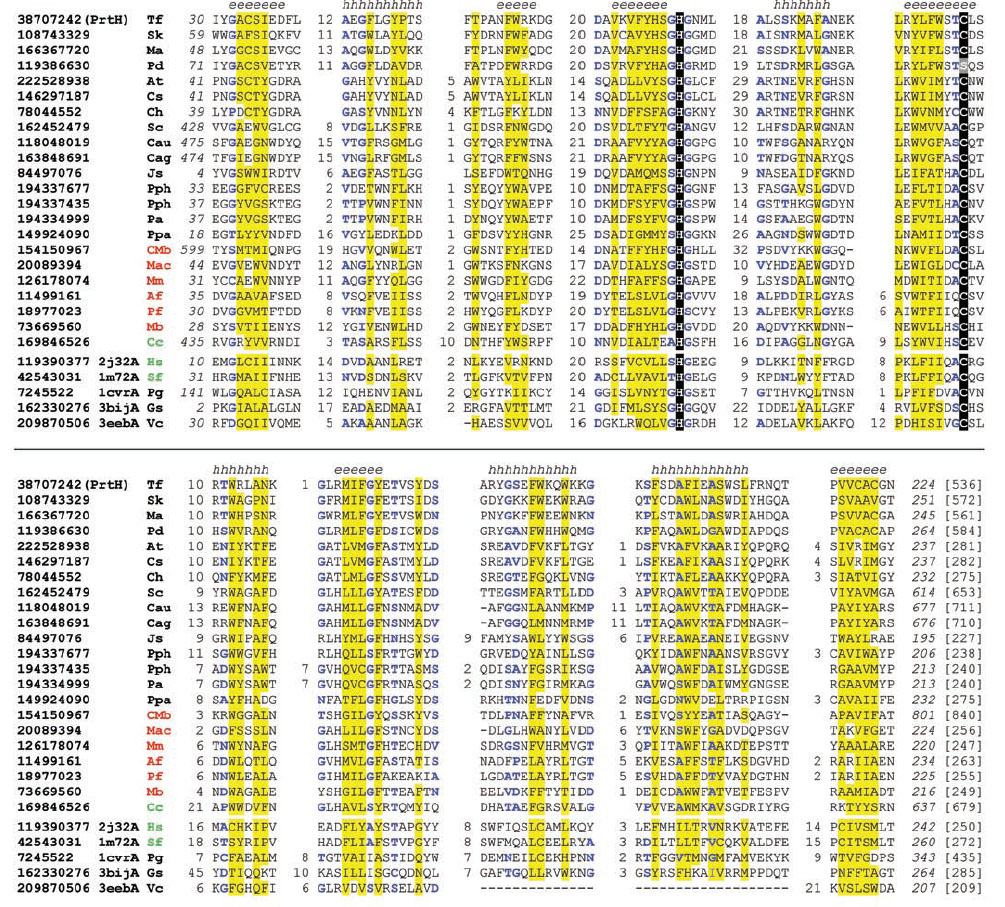
Multiple sequence alignment of PrtH homologs and several structures of caspase-like fold. Tannerella forsythia is a bacterial pathogen involved in periodontal disease. A cysteine protease PrtH has been characterized in this bacterium as a virulence factor. PrtH has the activity of detaching adherent cells from substratum, and the level of PrtH is associated with periodontal attachment loss. No reports exist on the structure, active site, and catalytic mechanism of PrtH. Using comparative sequence and structural analyses, we have identified homologs of PrtH in a number of bacterial and archaeal species. PrtH was found to be remotely related to caspases and other proteases with a caspase-like fold, such as gingipains from another periodontal pathogen Porphyromonas gingivalis. Our results offer structural and mechanistic insights into PrtH and its homologs, and help classification of this protease family. This alignment was made using PROMALS3D followed by manual adjustment. Catalytic histidines and cysteines are shaded in black. Non-polar residues in positions with mainly hydrophobic residues are shaded in yellow. Small residues (G, A, S, C, T, N, D, V and P) in positions with mainly small residues are colored blue. Starting and ending residues numbers are shown in italic numbers and sequence lengths are shown in brackets. Insertion regions in the alignment are replaced by the numbers of residues. Consensus secondary structure predictions are shown above the alignment (h: α-helix; e: β-strand). The proteins are identified by their NCBI gene identification (gi) numbers, followed by the species name abbreviations. The last five sequences have known structures with a caspase-like fold. Their Protein Data Bank ids and chain ids are shown after the gi numbers (2j32, caspase 3; 1m72, caspase 1; 1cvr, gingipain; 3bij, a bacterial caspase homolog; 3eeb, cysteine protease domain from V. cholerae RTX toxin). Species name abbreviations are: Af, Archaeoglobus fulgidus; At, Anaerocellum thermophilum; Cag, Chloroflexus aggregans; Cau, Chloroflexus aurantiacus; Cc, Coprinopsis cinerea; Ch, Carboxydothermus hydrogenoformans; CMb, Candidatus Methanoregula boonei; Cs, Caldicellulosiruptor saccharolyticus; Gs, Geobacter sulfurreducens; Hs, Homo sapiens; Js, Janibacter sp.; Mac, Methanosarcina acetivorans; Ma, Microcystis aeruginosa; Mb, Methanosarcina barkeri; Mm, Methanoculleus marisnigri; Pa, Prosthecochloris aestuarii; Pd, Paracoccus denitrificans; Pf, Pyrococcus furiosus; Pg, Porphyromonas gingivalis; Pph, Pelodictyon phaeoclathratiforme; Ppa, Plesiocystis pacifica; Sc, Sorangium cellulosum; Sf, Spodoptera frugiperda; Sk, Streptomyces kanamyceticus; Tf, Tannerella forsythia; Vc, Vibrio cholerae. Bacterial, archaeal, and eukaryotic species name abbreviations are shown in black, red and green respectively.