d1a05a_d1dgsa3
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1a05_Ai
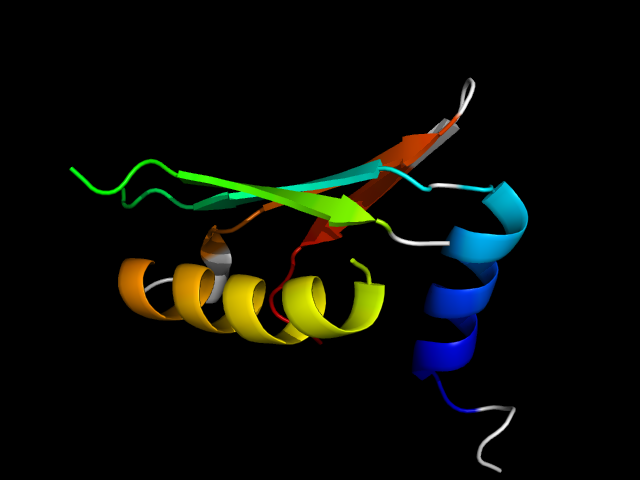 |
d1a05a_ | A82-A110,A126-A136,A241-A280 | Alpha and beta proteins (a/b) | Isocitrate/Isopropylmalate dehydrogenase-like | Isocitrate/Isopropylmalate dehydrogenase-like | Dimeric isocitrate & isopropylmalate dehydrogenases | 3-isopropylmalate dehydrogenase, IPMDH |
1dgs_Ac
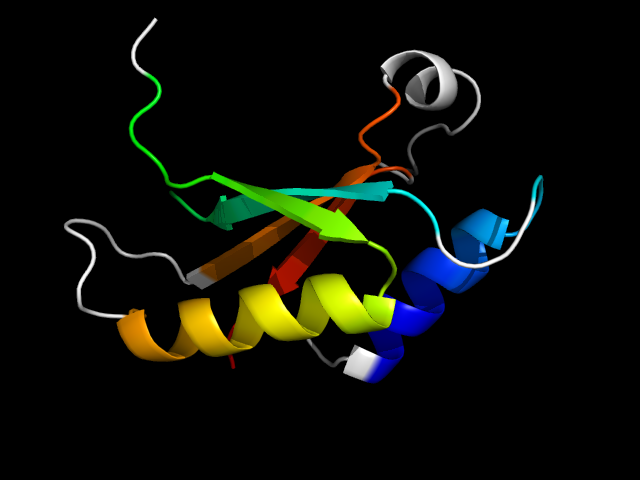 |
d1dgsa3 | A89-A118,A249-A259,A260-A314 | Alpha and beta proteins (a+b) | ATP-grasp | DNA ligase/mRNA capping enzyme, catalytic domain | Adenylation domain of NAD+-dependent DNA ligase | Adenylation domain of NAD+-dependent DNA ligase
|
Justification for analogy
In 1a05, motif occurs at the interface of the two duplicates.
In 1dgs, the motif is a circularly permuted reverse-ferredoxin core.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai ppakRPEQGLLRLRKGLD--lYANLRPAQIF--DVDILVVREltGNMFGDILSDEASQLTgs----igMLPSASLGe-----------graMYEPIHGS
1dgs_Ac -ftyEEVLAFEERLEREAeapSLYTVEHKVDfpVEHCYEKAL--GAEGVEEVYRRGLAQRhalpfeadGVVLKLDDltlwgelgytaraprFALAYKFP
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai ppakrpEQGLLRLRKGL-------dlYANLRPaqifdvdILVVRELT---gnmFGDILSDEASQLTGSIgmLPSASLG---------------egRAMYEPIHgs
1dgs_Ac ----ftYEEVLAFEERLereaeapslYTVEHK--vdfpvEHCYEKALgaegveEVYRRGLAQRHALPFE-aDGVVLKLddltlwgelgytaraprFALAYKFP--
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai ---PPAKRPE-QGLLRLRKGLD--LYANLRPAQIF---DVDILVVRELTGNMFGDILSDEASQ-LTG-----SIGMLPSASLGEGRA-----------MYEPIHGS
1dgs_Ac FTY-----EEVLAFEERLEREAEAPSLYTVEHKVDFPV-EHCYEKALG--AEGVEE-VYRRGLAQRHALPFEADG-VVLKLDDLTLWGELGYTARAPRFALAYKFP
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai PPAKR----PEQGLLRLRKGLDLY----ANLRPAQIFDVD--------ILVVRELTGNMFGDILSDEASQLTGSIGM----------LPSASLGEGRAM-----------------YEPIHGS---
1dgs_Ac -----FTYEEVLAFEERLEREA--EAPSLYTVEH------KVDFPVEHCYEKAL---GAEGVEEVYRRGLAQ-----RHALPFEADGVVLKLD------DLTLWGELGYTARAPRFALAY---KFP

