d1a05a_d1j71a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1a05_Ai
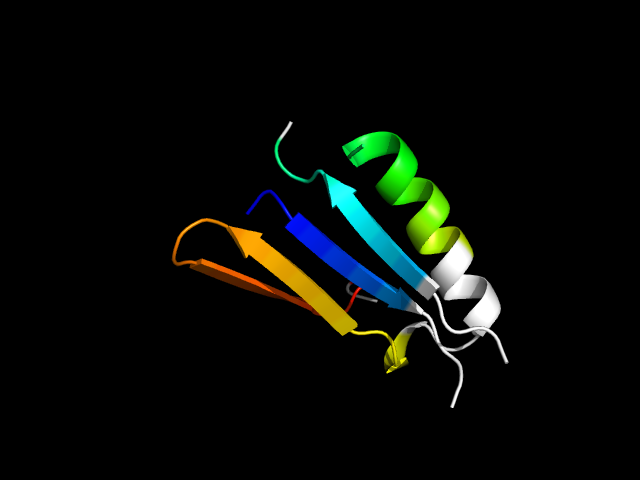 |
d1a05a_ | A100-A110,A126-A137,A241-A281 | Alpha and beta proteins (a/b) | Isocitrate/Isopropylmalate dehydrogenase-like | Isocitrate/Isopropylmalate dehydrogenase-like | Dimeric isocitrate & isopropylmalate dehydrogenases | 3-isopropylmalate dehydrogenase, IPMDH |
1j71_Ac
 |
d1j71a_ | A192-A205,A206-A221,A222-A334 | All beta proteins | Acid proteases | Acid proteases | Pepsin-like | Acid protease
|
Justification for analogy
The motif in 1a05: motif occurs at the interface of the two duplicates.
The motif in 1j71: a core motif. Alternatively, this can be regarded as an interface motif because it uses corresponding elements from the two duplicates.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai -------LYANLRPaqifdvdiLVVRELTg---------------GNMFGDILSDEasqltg--------siGMLPSASLGEG-RAMYEPIHGsa--------------------------------------------------------
1j71_Ac svelrvhLGSINFD--------GTSVSTNadvvldsgttityfsqSTADKFARIVGatwdsrneiyrlpscdLSGDAVFNFDQgVKITVPLSElilkdsdssicyfgisrndanilgdnflrrayivydlddktislaqvkytsssdisal
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai ------lYANLRPaqifdvdILVVRELTG----------GNMFGDILSDEASQL------------tgsIGMLPSASLGEgRAMYEP---iHGSA------------------------------------------------------
1j71_Ac svelrvhLGSINF-----dgTSVSTNADVvldsgttityFSQSTADKFARIVGAtwdsrneiyrlpscdLSGDAVFNFDQ-GVKITVplseLILKdsdssicyfgisrndanilgdnflrrayivydlddktislaqvkytsssdisal
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai -------LYANLRPAQIFDVDILVVRELTG-----------GNM-FGDILSDEASQL-----------TGS-IGMLPSASLG-EGRAMYEPI--HGSA------------------------------------------------------
1j71_Ac SVELRVHLGSINFDG-------TSVSTNADVVLDSGTTITY-FSQSTADKFARIVGATWDSRNEIYRL-PSCDLSGDAVFNFDQGVKITVPLSELILKDSDSSICYFGISRNDANILGDNFLRRAYIVYDLDDKTISLAQVKYTSSSDISAL
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai L--------YANLRPAQIFDVD--ILVVRELTGGN--------------MFGDILSDEASQLTGSIG-----------------MLPSASLGEG-RAMYEPIHGSA-----------------------------------------------------------
1j71_Ac -SVELRVHLGSINF--------DGTSVSTNA----DVVLDSGTTITYFSQSTADKFARIVG------ATWDSRNEIYRLPSCDLSGDAVFNFDQGVKITVPLS---ELILKDSDSSICYFGISRNDANILGDNFLRRAYIVYDLDDKTISLAQVKYTSSSDISAL

