d1a05a_d1rblm_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1a05_Ai
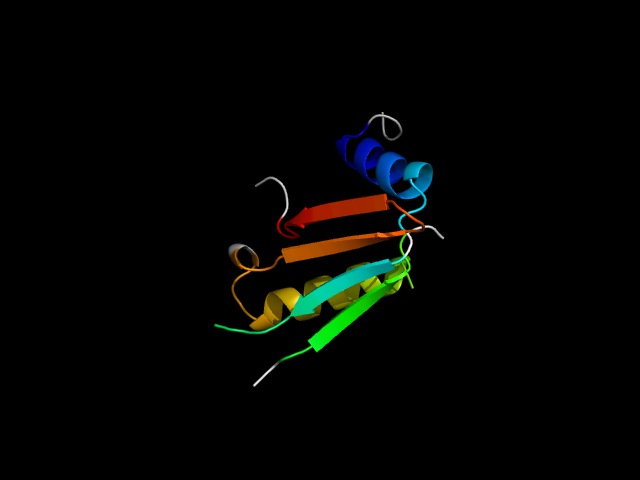 |
d1a05a_ | A82-A110,A126-A137,A241-A281 | Alpha and beta proteins (a/b) | Isocitrate/Isopropylmalate dehydrogenase-like | Isocitrate/Isopropylmalate dehydrogenase-like | Dimeric isocitrate & isopropylmalate dehydrogenases | 3-isopropylmalate dehydrogenase, IPMDH |
1rbl_Mc
 |
d1rblm_ | M2-M51,M64-M77,M78-M122 | Alpha and beta proteins (a+b) | RuBisCO, small subunit | RuBisCO, small subunit | RuBisCO, small subunit | Ribulose 1,5-bisphosphate carboxylase-oxygenase
|
Justification for analogy
The motif in 1a05: motif occurs at the interface of the two duplicates. circularly permuted reverse-ferredoxin.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai ------------------ppakRPEQGLLRLRKGLDLyANLRPAQIF-----dvDILV--VRELtg-GNMFGDILSDEASQLTGSIgMLPSASLGE--GRA-MYEPIHgsa-
1rbl_Mc smktlpkerrfetfsylpplsdRQIAAQIEYMIEQGF-HPLIEFNEHsnpeefyWTMWklPLFAc-aAPQQVLDEVRECRSEYGDC-YIRVAGFDNikECQtSSFIVHrpgr
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai ------------------ppakRPEQGLLRLRKGLDLyANLRP-------aqifdvdiLVVRELTGgnmfgDILSDEASQLTGSIGmlpSASLGEG---rAMYEPIH-gsa
1rbl_Mc smktlpkerrfetfsylpplsdRQIAAQIEYMIEQGF-HPLIEfnehsnpeefywtmwKLPLFACAapqqvLDEVRECRSEYGDCY-irVAGFDNIkecqTSSFIVHrpgr
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai ----------------------PPAKRPE-QGL-LRLR-KGLDLYANLRPAQIF--------DVDILV-VR-ELTGGNMFGDILSDEASQLTGSIGMLPSASLG-EG-R-AMYEPIH----GSA
1rbl_Mc SMKTLPKERRFETFSYLPPLSD-----RQIAAQIE-YMIE-QGF-HPLIEFNEHSNPEEFYW--TMWKLPLFAC-AAPQQVLDEVRECRSEYGDC-YIRVAGFDNIKECQTSSFIVHRPGR---
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a05_Ai PPAKR-----------------------PEQGLLRLRKGLDLYANLRPAQIFDVDILVVRELTGG--------------------NMFGDILSDEASQLTGSIG--MLPSASLGEGRA------MYEPIHGSA-----
1rbl_Mc -----SMKTLPKERRFETFSYLPPLSDRQIAAQIEYMIEQGF-HPLIEFNEH-------------SNPEEFYWTMWKLPLFACAAPQQVLDEVRECRSEYG---DCYIRVAGFDN---IKECQTSSFIV----HRPGR

