d1a2za_d1ghha_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1a2z_Ah
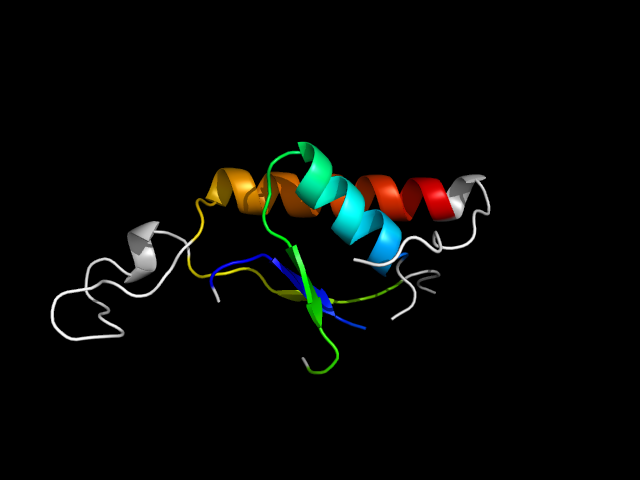 |
d1a2za_ | A74-A81,A114-A138,A159-A220 | Alpha and beta proteins (a/b) | Phosphorylase/hydrolase-like | Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) | Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) | Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
1ghh_Ac
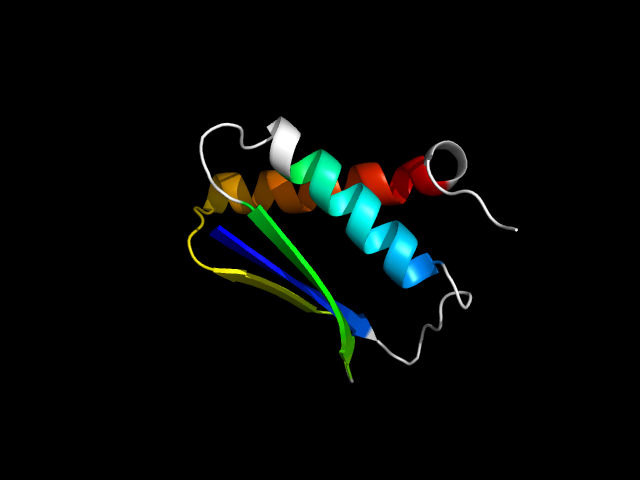 |
d1ghha_ | A1-A12,A13-A46,A47-A81 | Alpha and beta proteins (a+b) | DNA damage-inducible protein DinI | DNA damage-inducible protein DinI | DNA damage-inducible protein DinI | DNA damage-inducible protein DinI
|
Justification for analogy
d1a2za_ hits d1nc3a_ (other superfamily) with Dali Z 13.3, and d2pth__ (othersuperfamily) hits d1nc3a_ with Z 9.5, thus d1a2za_ and d2pth__
are homologs (the Z score between these two is 7.6, less than 9). In addition, the superposition of 1a2z and 2pth has some details suggesting
homology between them. Compare d1a2za_ and d2pth__, one strand and one helix in the motif are insertions. So this is a hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a2z_Ah ySNITVER------atlPVRAITKTLRDNG--------IPATISYSayplkAGFIHVPYtpdqvvnkffllgkntpsMCLEAEIKAIELAVKVSLDYlekdrddikipl
1ghh_Ac -MRIEVTIaktsplpagAIDALAGELSRRIqyafpdneGHVSVRYAa----ANNLSVIG------------------ATKEDKQRISEILQETWESAddwfvse-----
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a2z_Ah ySNITVER----------atlPVRAITKTLRDNG---iPATISYSAyplkaGFIHVPytpdqvvnkffllgkntpSMCLEAEIKAIELAVKVSLDYleKDRDdikipl
1ghh_Ac -MRIEVTIaktsplpagaidaLAGELSRRIQYAFpdneGHVSVRYA---aaNNLSVI------------------GATKEDKQRISEILQETWESA--DDWF---vse
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a2z_Ah -YSNITVE-R--ATL--PVRAITKTLRDNG--------IPATISYSAYPLKAGFIHVPY--TPD-QVVNKFFLLGKNTPSMCLEAEIKAIELAVKVSLD-YLEKDRDDIKIPL
1ghh_Ac M-RIEVTIAKTSPLPAGAIDALAGELSRRIQYAFPDNEGHVSVRYAA------ANNLSVIGATKE---------------DKQRISEILQETWESA-DDWF--V-SE------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a2z_Ah YS-NITVERATLP---------------VRAITKTLRDNG-----IPATISYSAYPLKAG----FIHVPYTPDQVVNKFFLLGKNTPSMCLEAEIKAIELAVKVSLDYLEKDRDDIKIPL------
1ghh_Ac --MRIEVT-----IAKTSPLPAGAIDALAGELSRRIQYA-FPDNEGHVSVRY--------AAANNLSVIG------------------ATKEDKQRISEILQETWESAD-----------DWFVSE

