d1a7j__d2if1__
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1a7j__h
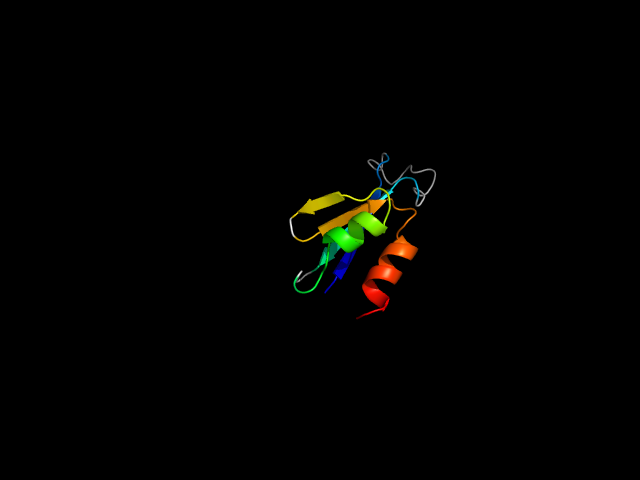 |
d1a7j__ | 203-279 | Alpha and beta proteins (a/b) | P-loop containing nucleoside triphosphate hydrolases | P-loop containing nucleoside triphosphate hydrolases | Phosphoribulokinase/pantothenate kinase | Phosphoribulokinase |
2if1__c
 |
d2if1__ | 1-126 | Alpha and beta proteins (a+b) | eIF1-like | eIF1-like | eIF1-like | Eukaryotic translation initiation factor eIF-1 (SUI1)
|
Justification for analogy
The motif in 1a7j: comparing d1a7j__ and 1uj2 (same SCOP family), three strands and one helix in the motif in 1a7j are absent in 1uj2. So
this is a hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a7j__h ------------------------------------------DINFQRVPVVdtsnpfiarwiptaDESVVVIRFrnpRGIDFPYLTSMIH-----GSWMSRa----NSIVVPGNKLDLAMQLILTPL------------
2if1__c mrgshhhhhhtdpmsaiqnlhsfdpfadaskgddllpagtedYIHIRIQQRNg-------------RKTLTTVQG-iaDDYDKKKLVKAFKkkfacNGTVIEhpeygEVIQLQGDQRKNICQFLVEIGlakddqlkvhgf
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a7j__h ------------------------------------------DINFQRVPVVDtsnpfiarwiptaDESVVVIRFrnPRGIDFPYLTSMIH-----GSWMSR----ANSIVVPGNKLDLAMQLILTPL------------
2if1__c mrgshhhhhhtdpmsaiqnlhsfdpfadaskgddllpagtedYIHIRIQQRNG-------------RKTLTTVQG-iADDYDKKKLVKAFKkkfacNGTVIEhpeyGEVIQLQGDQRKNICQFLVEIGlakddqlkvhgf
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a7j__h ------------------------------------------DINFQRVPVVDTSNPFIARWIPTADESVVVIRFRNPRGIDFPYLTSMIH-----GSWMSR----ANSIVVPGNKLDLAMQLILT-P--L---------
2if1__c MRGSHHHHHHTDPMSAIQNLHSFDPFADASKGDDLLPAGTEDYIHIRIQQRN-----------G--RKTLTTVQG-IADDYDKKKLVKAFKKKFACNGTVIEHPEYGEVIQLQGDQRKNICQFLVEIGLAKDDQLKVHGF
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1a7j__h ------------------------------------------DINFQRVPVVDTSNPFIARWIPTAD--ESVVVIRFRNPR-GIDFPYLTSMIH------GSWMSR-----ANSIVVPG-NKLDLAMQLILTPL-------------
2if1__c MRGSHHHHHHTDPMSAIQNLHSFDPFADASKGDDLLPAGTEDYIHIRIQQRN---------------GRKTLTTVQ--GIADDYDKKKLVKAF-KKKFACNGTVI-EHPEYGEVIQLQ-GDQRKNICQFLVEI-GLAKDDQLKVHGF

