d1aa7a_d1b68a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1aa7_Ai
 |
d1aa7a_ | A51-A88,A120-A158 | All alpha proteins | Influenza virus matrix protein M1 | Influenza virus matrix protein M1 | Influenza virus matrix protein M1 | Influenza virus matrix protein M1 |
1b68_Ac
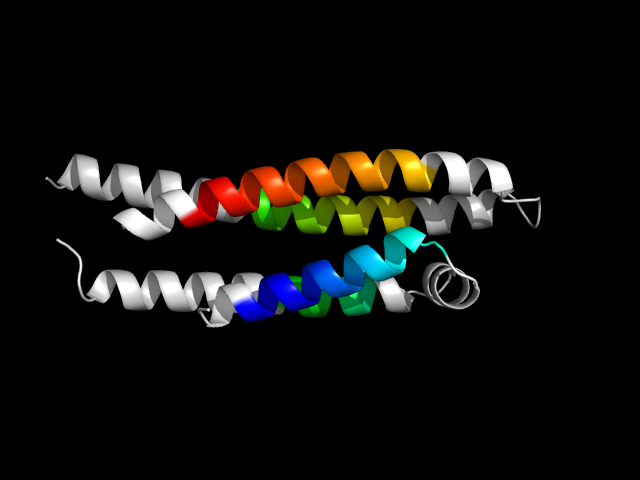 |
d1b68a_ | A23-A85,A86-A162 | All alpha proteins | Four-helical up-and-down bundle | Apolipoprotein | Apolipoprotein | Apolipoprotein E
|
Justification for analogy
The motif in 1aa7 uses helices from the two different domains and the linker helix, and is an interface motif.
Although the first domain in 1aa7_A is also a 4-helix up-and-down bundle, this bundle cannot be homologous to the 4-helix
bundle in 1b68, because they are mirror images, i.e. superimpose helix 1 and 2 in 1b68 and 1aa7, helix 3 and 4 are on
different sides of the plane formed by helix 1 and 2.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai ---ilSPLTKGILGFVFTLTVpser-----glqrRRFVQNALNgng--------------------------------SAGALASCMGLIYNRmg---------------avTTEVAFGLVCATCEQIADSQ------
1b68_Ac gqrweLALGRFWDYLRWVQTLseqvqeellssqvTQELRALMDetmkelkaykseleeqltaeetrarlskelqaaqaRLGADMEDVRGRLVQyrgevqamlgqsteelrvrLASHLRKLRKRLLRDADDLQkrlavy
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai ---iLSPLTKGILGFVFTLTVPS-ERGL----QRRRFVQNALN--------------------------------gngSAGALASCMGLIYNRM---------------gaVTTEVAFGLVCATCEQIADSQ------
1b68_Ac gqrwELALGRFWDYLRWVQTLSEqVQEEllssQVTQELRALMDetmkelkaykseleeqltaeetrarlskelqaaqaRLGADMEDVRGRLVQYrgevqamlgqsteelrvRLASHLRKLRKRLLRDADDLQkrlavy
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai -ILSPLTKGILGFVFTLTV--PS------E--R-----G--LQRR--RFVQNALN---------------------GNGSAGALASCMGLIYNR-MGA--------------------------V-T-TEVAFGLVCATCEQIADSQ
1b68_Ac G-Q---RW-ELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELRALMDETMKELKAYKSELEEQLTAEETRARLSK---ELQAAQARLGADMEDVRGRLVQYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVY-
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai ILS------PLTKGILGFVFTLTVPSERGLQRR---------------RFVQNALNGNG------------------------------------SAGALASCMGLIYNRMGAVT---------------------TEVAFGLVCATCEQIADSQ------
1b68_Ac ---GQRWELALGRFWDYLRWVQT----------LSEQVQEELLSSQVTQELRALM----DETMKELKAYKSELEEQLTAEETRARLSKELQAAQARLGADMEDVRGRLV------QYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVY

