d1aa7a_d1qkra_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1aa7_Ai
 |
d1aa7a_ | A1-A16,A51-A88,A120-A158 | All alpha proteins | Influenza virus matrix protein M1 | Influenza virus matrix protein M1 | Influenza virus matrix protein M1 | Influenza virus matrix protein M1 |
1qkr_Ah
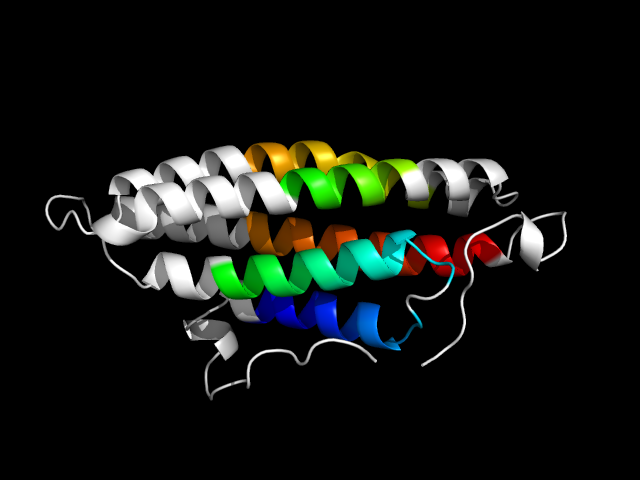 |
d1qkra_ | A881-A914,A915-A973,A974-A1061 | All alpha proteins | Four-helical up-and-down bundle | alpha-catenin/vinculin | alpha-catenin/vinculin | Vinculin
|
Justification for analogy
The motif in 1aa7 uses helices from the two different domains and the linker helix, and is an interface motif.
Although the first domain in 1aa7_A is also a 4-helix up-and-down bundle, this bundle cannot be homologous to the 4-helix bundle in 1qkr,
because they are mirror images.
Note that in 1qkr paper, the authors mentioned that "five-helix bundles are uncommon,.." in page 606.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai ------------------mSLLTEVETYVLSIIpiLSPLTKGILGFVFTLTVp--------------serglqrRRFVQNALNgng-------------SAGALASCMGLIYNRmga---------------------------vTTEVAFGLVCATCEQIADSQ----------------
1qkr_Ah kdeefpeqkageainqpmmMAARQLHDEARKWSskGNDIIAAAKRMALLMAEmsrlvrggsgnkraliqcakdiAKASDEVTRlakevakqctdkrirtNLLQVCERIPTISTQlkilstvkatmlgrtnisdeeseqatemlvhNAQNLMQSVKETVREAEAASikirtdagftlrwvrk
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai ----------MSLLTEV-ETYVLSIIP--------------ilSPLTKGILGFVFTLTVpsERGLQRR--RFVQNALN---------------------------gngSAGALASCMGLIYNR-------------mgaVTTEVAFGLVCATCEQIADSQ-----------------------
1qkr_Ah kdeefpeqkaGEAINQPmMMAARQLHDearkwsskgndiiaaaKRMALLMAEMSRLVRG--GSGNKRAliQCAKDIAKasdevtrlakevakqctdkrirtnllqvceRIPTISTQLKILSTVkatmlgrtnisdeeseQATEMLVHNAQNLMQSVKETVreaeaasikirtdagftlrwvrk
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai ---------------------------------------------------------------------M--SLLTEVETYVLSIIP-IL-------SPLTKGI-LGFVFTLTVP---------------------------SERGLQRRRFVQNALNGNGSAGALASCMGLIYNRMGA-VTTEVAFGLVCATCEQIADS-Q----------------
1qkr_Ah KDEEFPEQKAGEAINQPMMMAARQLHDEARKWSSKGNDIIAAAKRMALLMAEMSRLVRGGSGNKRALIQCAKDIAKASDEVTRLAKEVAKQCTDKRIRTNLLQVCE-RIPTISTQLKILSTVKATMLGRTNISDEESEQATE------------------------------------MLVHNAQNLMQSVKETVREAEAASIKIRTDAGFTLRWVRK
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1aa7_Ai MSLLT----------------EVETYVLSIIPILS-----------------PLTKGILGFVFTLTVPSERGLQRR---RFVQNALNGNGS-----------------------------------AGALASCMGLIYNRMGAVT-------------------TEVAFGLVCATCEQIADSQ-----------------------
1qkr_Ah -----KDEEFPEQKAGEAINQPMMMAARQLHD---EARKWSSKGNDIIAAAKRMALLMAEMSRLVRGGS-------GNKRALIQCAK----DIAKASDEVTRLAKEVAKQCTDKRIRTNLLQVCERIPTISTQLKILST------VKATMLGRTNISDEESEQATEMLVHNAQNLMQSVKETVREAEAASIKIRTDAGFTLRWVRK

