d1ac5__d1jroa3
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1ac5__h
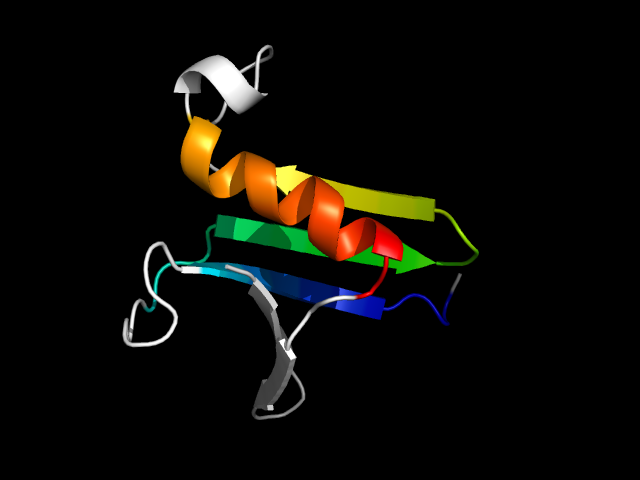 |
d1ac5__ | 407-483 | Alpha and beta proteins (a/b) | alpha/beta-Hydrolases | alpha/beta-Hydrolases | Serine carboxypeptidase-like | Serine carboxypeptidase II |
1jro_Ac
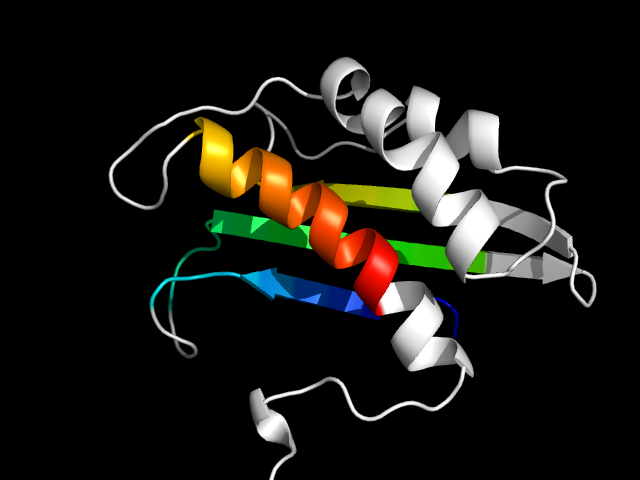 |
d1jroa3 | A346-A462 | Alpha and beta proteins (a+b) | CO dehydrogenase flavoprotein C-domain-like | CO dehydrogenase flavoprotein C-terminal domain-like | CO dehydrogenase flavoprotein C-terminal domain-like | Xanthine dehydrogenase chain A, domain 4
|
Justification for analogy
The motif in 1ac5: four strands are insertions to the Rossmann core, while one strand and the helix belong to the Rossmann core. Hybrid
motif, but the b-a-b connection in this motif is left-handed.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ac5__h sDDAVSFDWIHkskstddsEEFSGYVKYDR---------NLTFVSVynash-----------------------------------mvpfdKSLVSRGIVDIYSNdvmiidnngknvmitt-
1jro_Ac -PGLRCYKLSKrf----dqDISAVCGCLNLtlkgskietARIAFGGmagvpkraaafeaaligqdfredtiaaalpllaqdftplsdmrasAAYRMNAAQAMALRyvrelsgeavavlevmp
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ac5__h sddAVSFDWIHKsKSTDDSEE-fSGYVKYDR-----NLTFVSVYNASH------------------------------MVPFD----------KSLVSRGIVDIYSND----vMIIDNngknvmitt
1jro_Ac ---PGLRCYKLS-KRFDQDISavCGCLNLTLkgskiETARIAFGGMAGvpkraaafeaaligqdfredtiaaalpllaQDFTPlsdmrasaayRMNAAQAMALRYVRElsgeaVAVLE------vmp
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ac5__h SDDAVSFDWIHKSKSTDDSE---E-FSGYVKYDR----N-LTFVSVYNAS-------------------------------HMVPFD----------KSLVSRGIVDIYSNDV-----MII-----DNNGKNVMITT
1jro_Ac ---PGLRCYKLS----KRFDQDISAVCGCLNLTLKGSKIETARIAFGGMAGVPKRAAAFEAALIGQDFREDTIAAALPLLAQDF-TPLSDMRASAAYRMNAAQAMALRY-VRELSGEAVAVLEVMP-----------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ac5__h SD-DAVSFDWIHKSKSTDDSE--EFSGYVKYDRN-----------LTFVSVYNASHMVPFDK-----------------------------------------------SLVSRGIVDIYSNDVMIIDNNGKNVMITT------------------
1jro_Ac --PGLRCYKL--SKRFD----QDISAVCGCLN--LTLKGSKIETARIAFG------------GMAGVPKRAAAFEAALIGQDFREDTIAAALPLLAQDFTPLSDMRASAAYRMNAAQAMAL-----------------RYVRELSGEAVAVLEVMP

