d1adja2d1drw_2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1adj_Ah
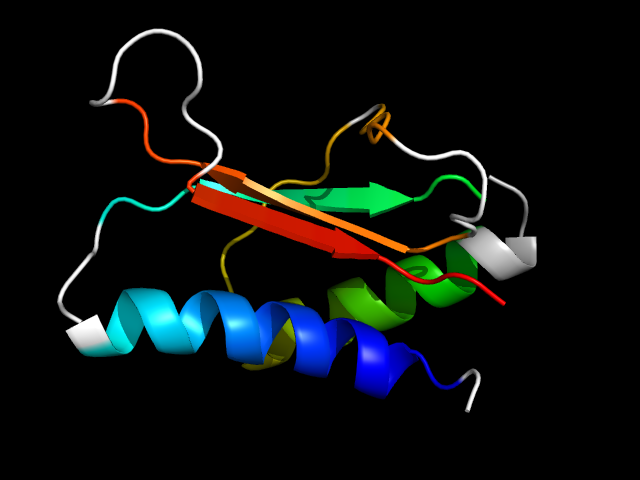 |
d1adja2 | A134-A167,A229-A289 | Alpha and beta proteins (a+b) | Class II aaRS and biotin synthetases | Class II aaRS and biotin synthetases | Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain | Histidyl-tRNA synthetase (HisRS) |
1drw__c
 |
d1drw_2 | 131-163,164-240 | Alpha and beta proteins (a+b) | Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain | Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain | Dihydrodipicolinate reductase-like | Dihydrodipicolinate reductase
|
Justification for analogy
The motif in 1adj: comparing d1adja2 and d1bia_3 (different family same superfamily), the helix and the second strand are insertion. hybrid. the
inserted beta-strand is short.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1adj_Ah seNPILDAEAVVLLYECLKElglrRLKVKLSSVG----lgeeaRAHLKEVERHLERLS---------------------VPYELEpALVRGldyyvRTAFEVHHEeigaqsaLGGGGRYD---
1drw__c -vGVNVMLKLLEKAAKVMGD----YTDIEIIEAHhrhk-vdapSGTALAMGEAIAHALdkdlkdcavysreghtgervpGTIGFA-TVRAGdi-vgEHTAMFADI-----geRLEITHKAssr
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1adj_Ah sENPILDAEAVVLLYECLkelglrRLKVKLSSVG---LGEEaRAHLKEVERHLERLS---------------------vPYELEPAlvrgldyyVRTAFEVHHeeigAQSAlGGGGRYD---
1drw__c -VGVNVMLKLLEKAAKVM----gdYTDIEIIEAHhrhKVDApSGTALAMGEAIAHALdkdlkdcavysreghtgervpgTIGFATV--ragdivGEHTAMFAD----IGER-LEITHKAssr
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1adj_Ah SENPILDAEAVVLLYECLKELGL-RRL-KVKLSSVGLGEEARAHLKEVERHLERLSV------P-------------------YELEPA----LVRGLDYYVRTAFEVHHEEIGAQSALGGGGRYD--
1drw__c V----GVNVM-LKLLEKAAK-VMGDYTDIEIIEAHHRHK--VDAPSGTALAMGEAIAHALDKDLKDCAVYSREGHTGERVPGTIGFATVRAGD-------IVGEHTAMFADIG---ERLEITHKASSR
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1adj_Ah SENPIL-DAEAVVLLYECLKELGLR----RLKVKLSSVGLGEEARAHLK--------EVERHLERLSV-----------------------------PYELEPALVRGLDYYVR-----TAFEVHHEEI-GAQSAL--GGGGRYD-
1drw__c ------VGVNVMLKLLEKAAKVM--GDYTDIEIIEAHH-----------RHKVDAPSGTALAMGEA--IAHALDKDLKDCAVYSREGHTGERVPGTIGFATVR-----------AGDIVGEHTAMFA--DIGER--LEITHKASSR

